Gene Comparison
The Gene Comparison app provides a heatmap visualization of average expression levels similar to the gene info app, but for multiple selected genes.
Feature Abundance

The text fields on the left-hand side are used to enter NCBI gene IDs, Ensembl IDs, or gene symbols. If the project comprises multiple species, the species of interest can be selected on the left-hand side using the Sample selector.
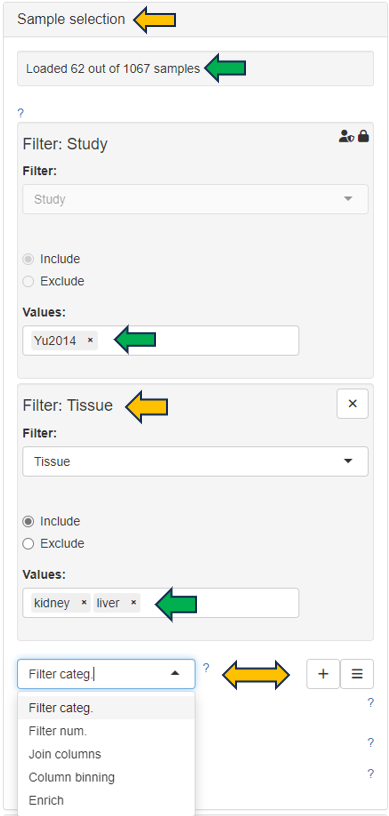
In case of a gene symbol, all selected species are searched. In case no species is selected, the default is used. You can also specify the species explicitly by adding the species code after the symbol separated by a blank. E.g. “abca1 hs” will find the human ABCA1 gene. Note that only genes which are present in the expression table can be found.
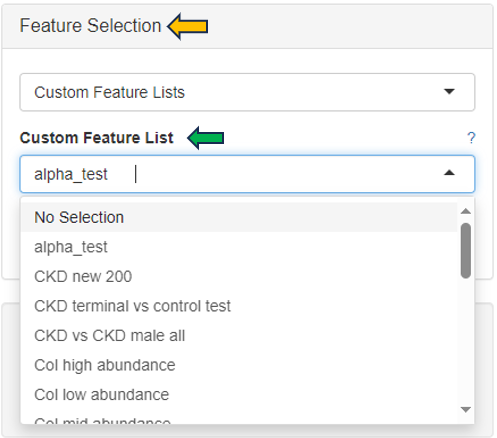
You can choose your feature list to only display the chosen features from the drop down menu under “Feature Selection”.
Using the “Group by” option, you can choose categories by which to group the expression data. Expression data will be averaged for each combination of chosen categories. Note that the order here (adjustable via drag & drop) also affects the order in which the categories are displayed in the plots / tables. If empty, the expression data will be grouped by ‘SampleID’, or in case of scRNA-Seq data by ‘Study’ and ‘scCluster’.

Table

The normalized feature abundance levels are averaged for the selected category values (for details about the interactive result table, see the description of the gene info app) and color-coded (dark green corresponds to high expression, light green or white to low expression).
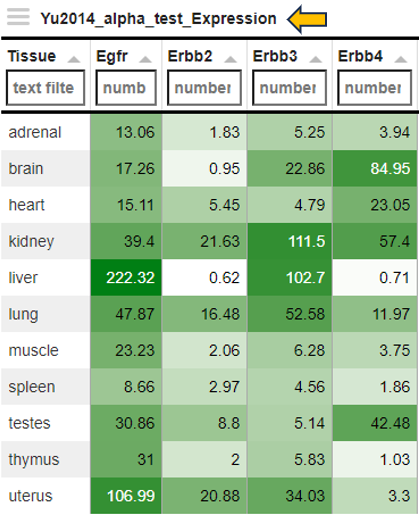
Heatmap

Display as a heatmap of only the selected features from the feature list. You can edit the heatmap with the “Options” presented to you above the diagram. The expression heatmap can be downloaded in the excel format using the given download option.
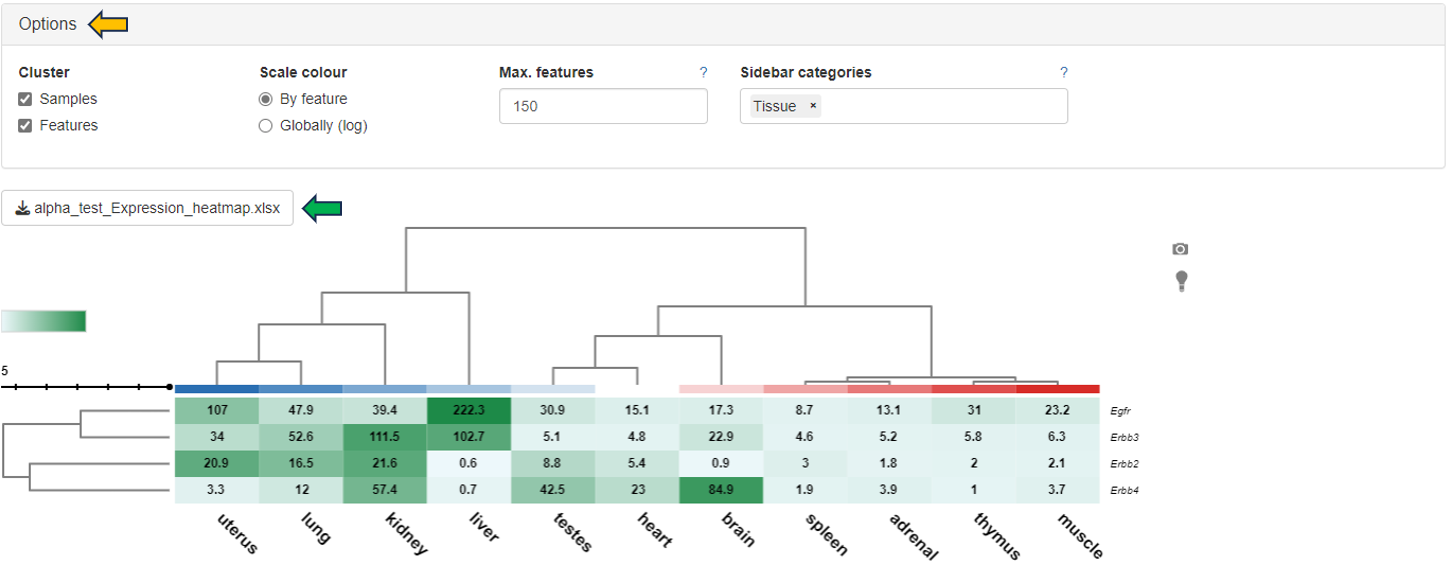
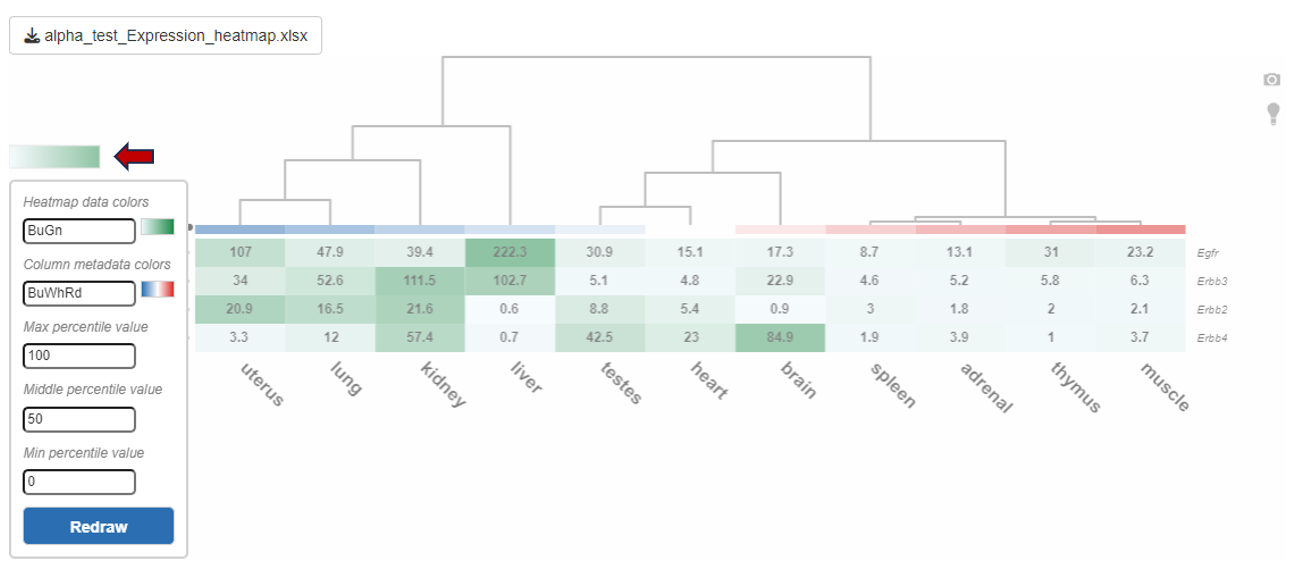
You can also edit the colors used for the depiction of heatmap using the “Color settings” option near the diagram.
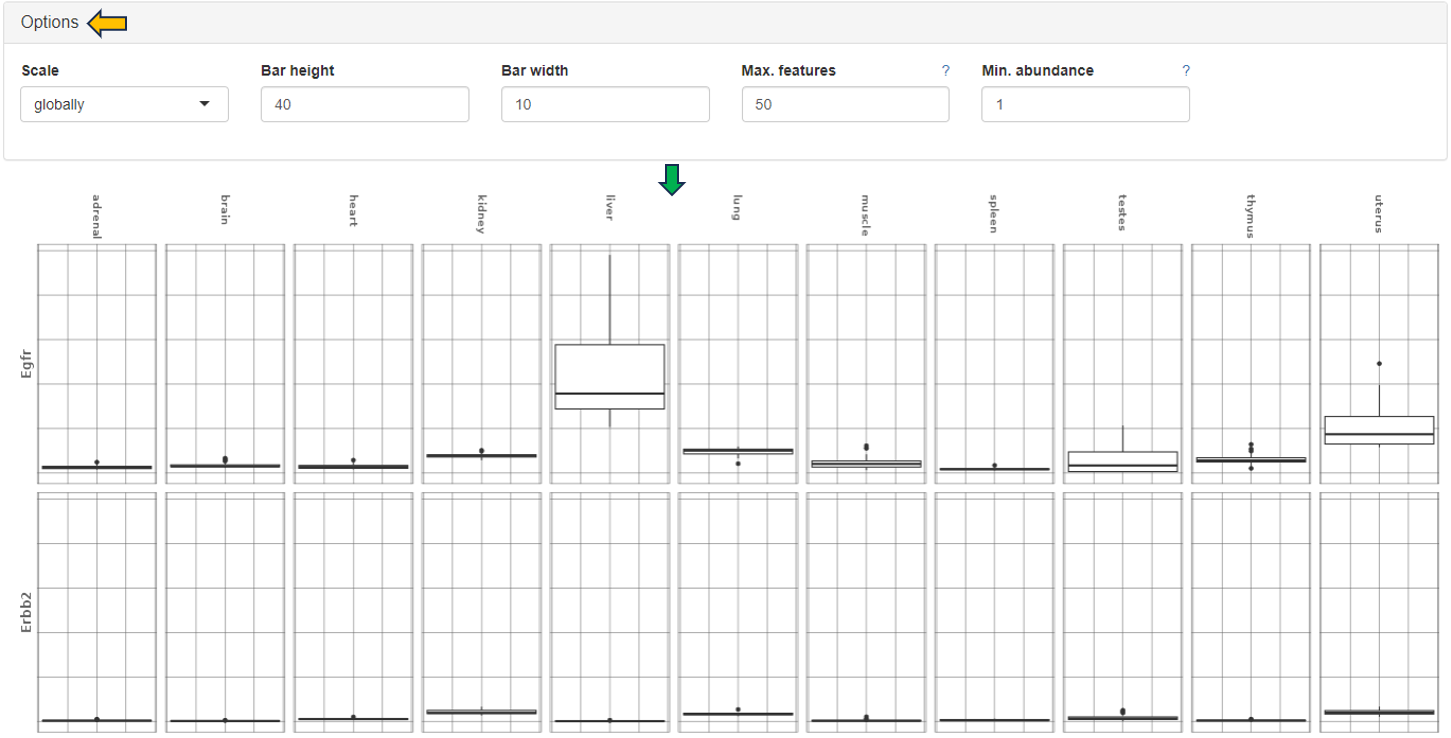
Barplots

This sub-panel allows the display of the expression data as a barplot.
Feature List
This panel provides a collection of features (e.g. genes or proteins) that can be used for further analysis. One can use the existing feature lists or create one’s own. You can create your own features list by adding features to this text panel.
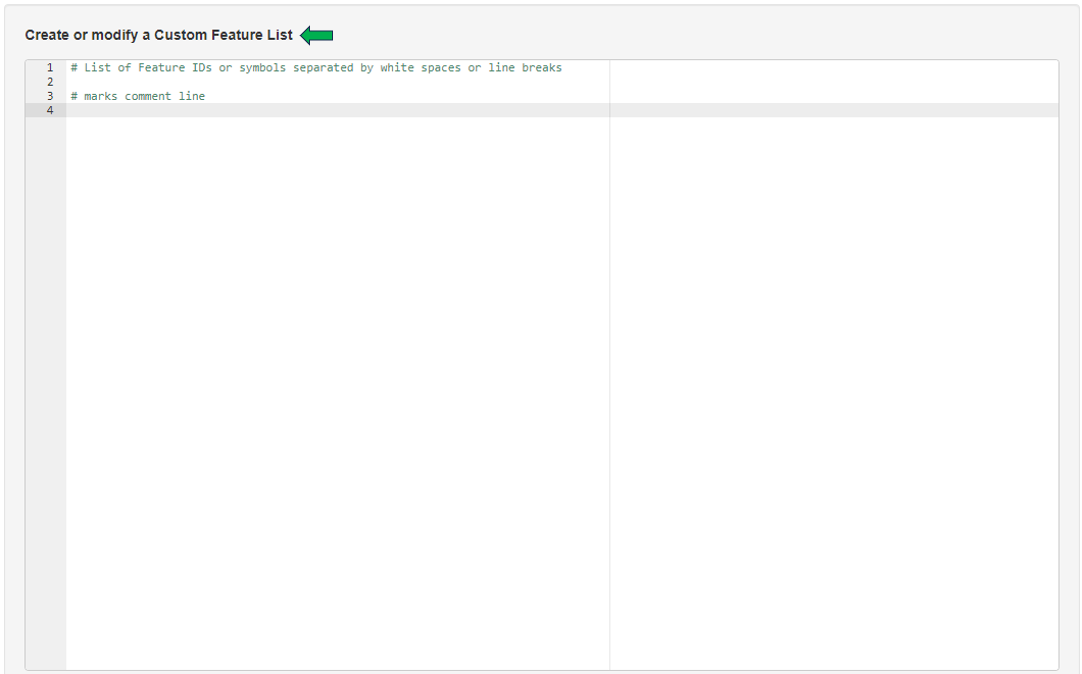
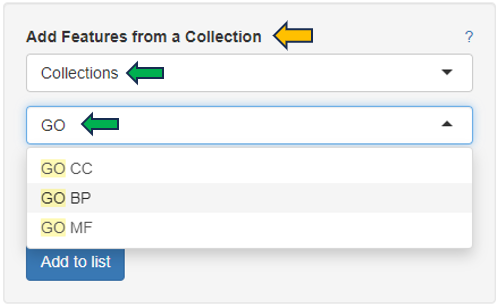
It is also possible to access lists that have already been saved, with the help of the “Features selector”. One possibility is to use the “Collection option” in the drop-down.Type GO into the tab below to see all the saved predefined lists from “GO (Gene ontology resource)” feature lists. These lists can be modified with the help of the above shown text panel and saved again to form a custom Feature List.
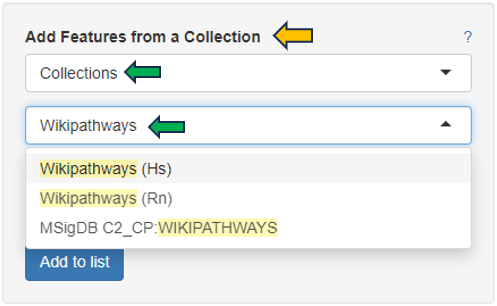
Similarly, another option is to use “Wikipathways”. Type Wikipathways to see all available Feature Lists. It is also possible to combine different lists and save them as one single list.
Another possibility is to access the Feature List of significant features from an already saved “Comparisons option”. Click the tab (under the Comparisons option) to look for all saved Comparisons table.
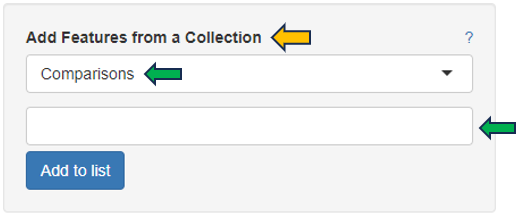
As soon as you click the tab, the following table pops up for you to select the features from the “Comparisons Selector”. You can then select from the various options available from both “Single Comparisons” tab and “Comparisons from Groups” tab. You can also search for the desired study or geneID in the given search box.
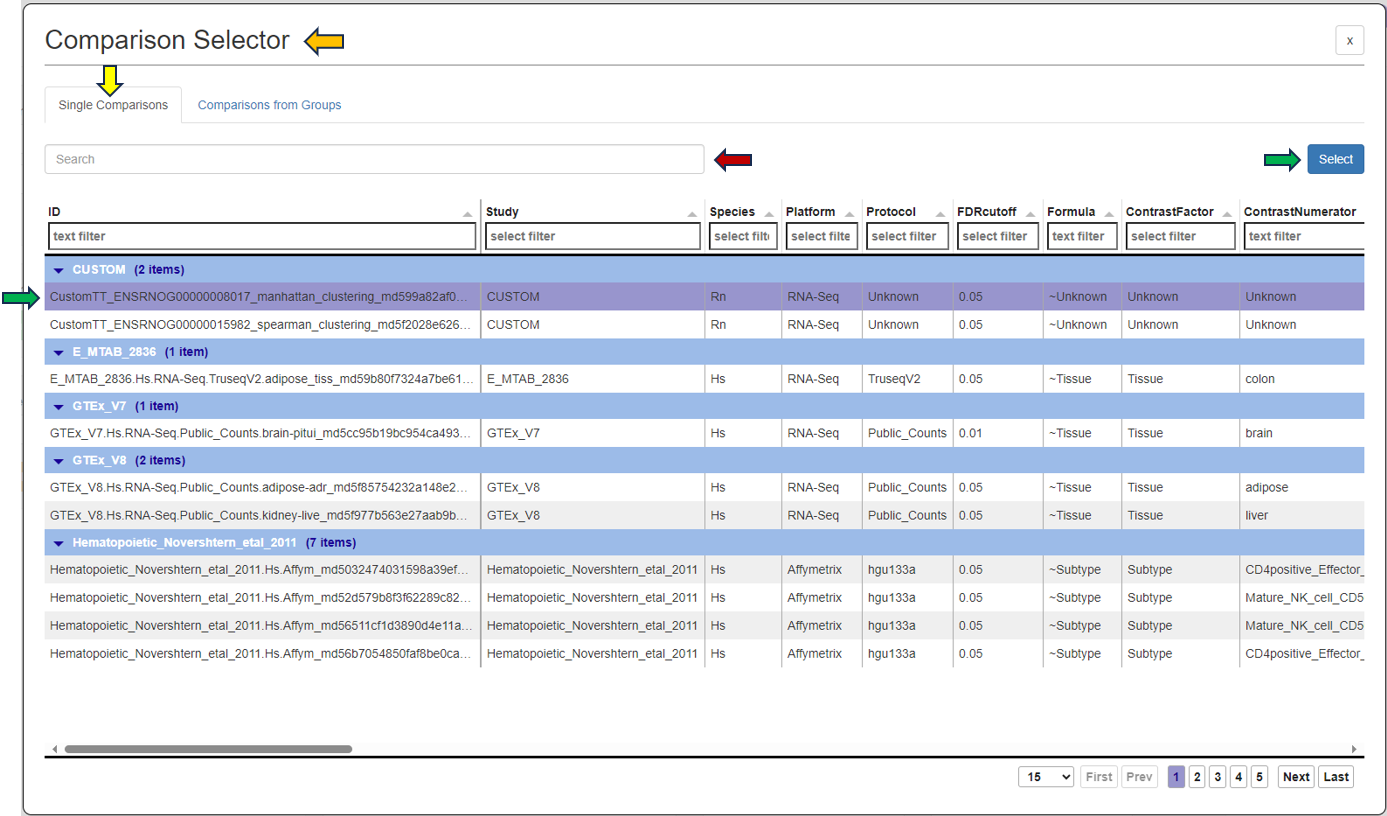
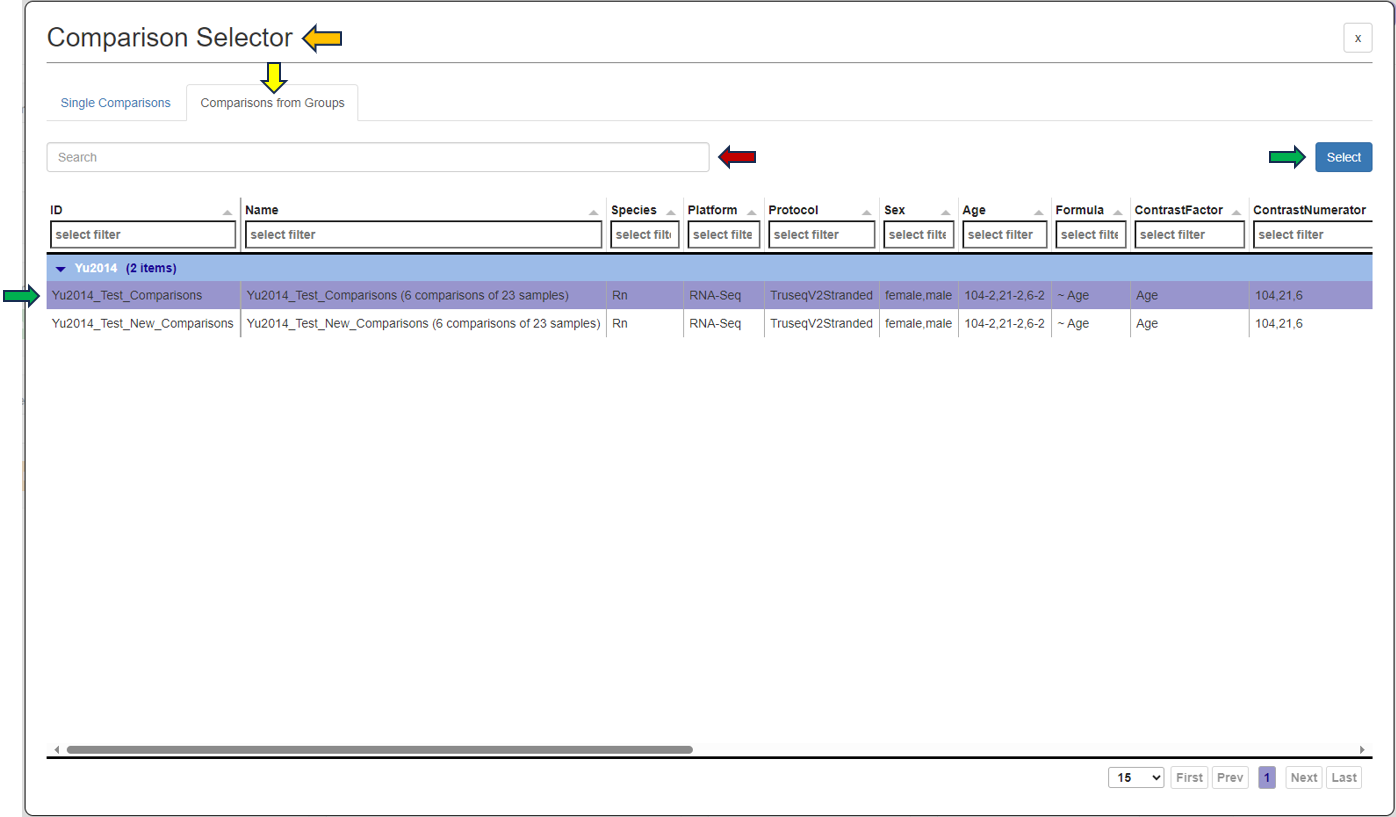
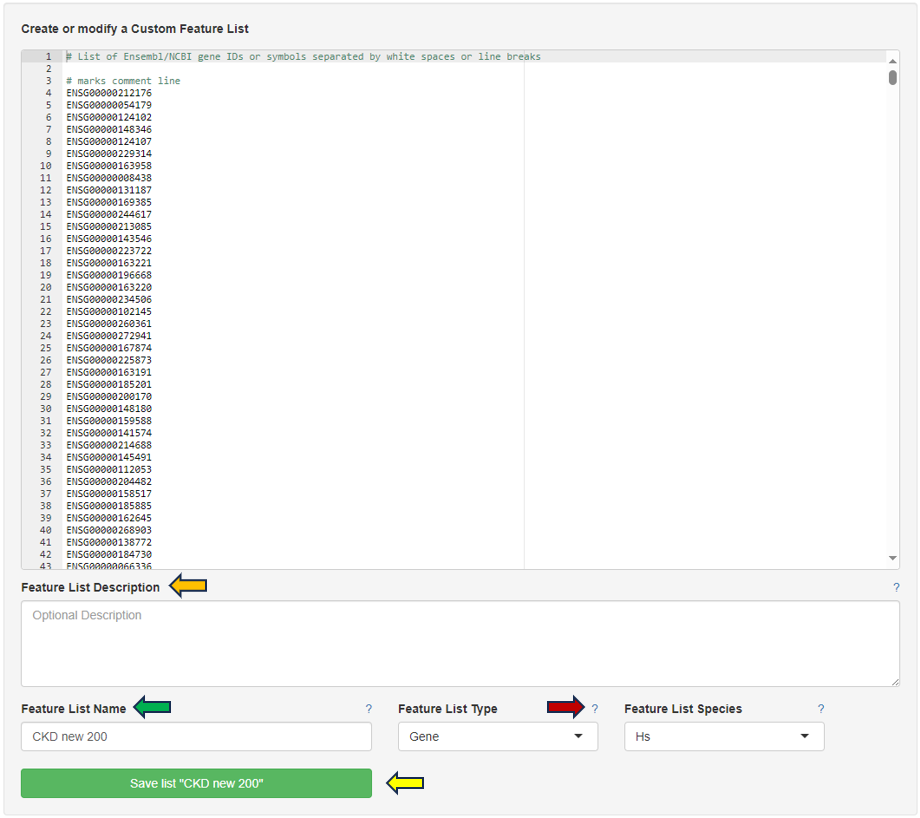
By clicking the question mark next to the “Feature List Type”, you will get a list of accepted IDs.You can then give the feature list a name and description in the “Feature List Name” and “Feature List Description” panel respectively, and save it to make it available to other apps.