Mapping between peptidomics and/or proteomics features
Since PanHunter is a true multi-omics platform, the challenge arises to match features or information from different entities. Especially for proteomics and peptidomics data this is of high relevance, as the measured intensities usually cannot be associated to a single protein, but only to a group of possible proteins. Such groups may only contain isoforms of the same canonical protein, but they can also contain completely independent proteins. Comparing the measurements of such protein groups (PGs) across different datasets is a common task in the data analysis.
IDs for proteins and peptide/PTMs
The whole procedure of mapping features between entities relies on well-defined IDs, that uniquely identify a feature. This can be for example the ID used from the “ensembl” database or from the ncbi database. For the identification of proteins, PanHunter uses the Uniprot ID. Protein groups on the other hand are then simply a list of such Uniprot IDs.
Peptides are not part of any of these databases, so PanHunter allows to use arbitrary IDs. The only requirement for them is that they are unique within a study. During the preprocessing the used software DiaNN, Maxquant or Spectronaut is identifying from which protein a peptide is coming from. Hence, each peptide gets associated to a protein group of possible proteins. In this process, also post-translational modifications (PTMs) are identified and the position and the type of the modified amino acid is retrieved. As the ID can be arbitrary, it is not guaranteed that it is valid across different studies. Hence, for this purpose PanHunter creates an internal ID. Currently, this internal ID consists out of the associated protein group, the modified amino acid and the location, but this might change in the future. Uniqueness is not a strict requirement for the internal ID, even if the probability for it is very high.
Mapping algorithms
As stated above, it is often required to match features from different entities. So, one dimension of the used algorithms are the types of the involved features, e.g a PTM/PTM, a PTM/PG or a PG/PG match. Apart from that, there are two algorithms per type available, namely the “exact matching” and an “explosion matching”. The details and the use-cases are explained below. Additionally, the workflows are also visualized by graphs. There, the columns used for a merge are colored in the same way. The FDR column is used here, as a placeholder for all additional columns that might be present.
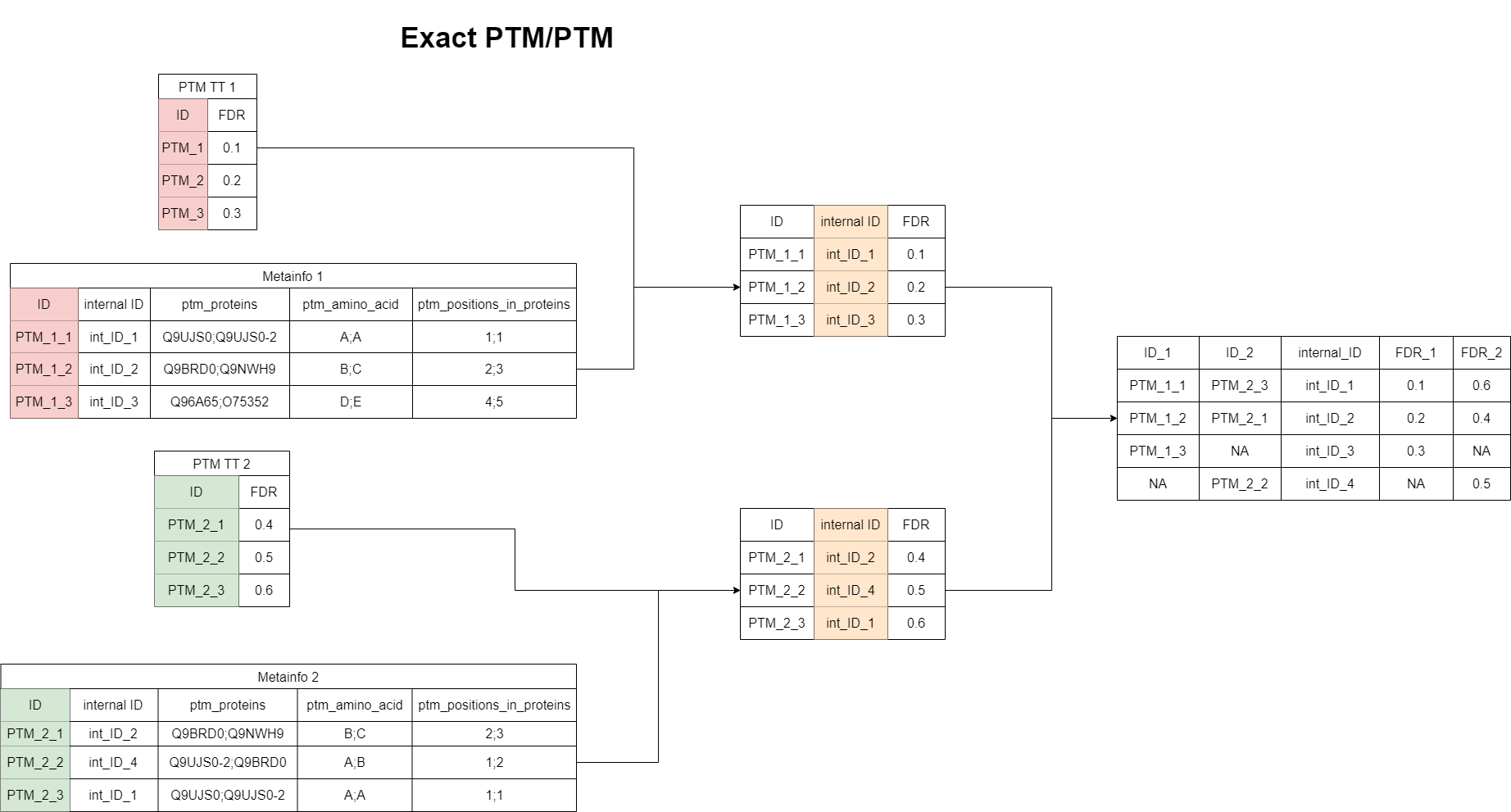
“Exact matching” for the PTM/PTM case
This case is the most simple one, as it is done based on the internal ID. Two PTM features are matched, if their internal IDs are fully identical. This means the same PG has been associated to them and all modifications are also identical. No isoform removal or any other modification of the associated protein groups are applied here. Therefore, this algorithms is quite strict, but the results are reliable in a sense, that the matched features are exactly identical in both datasets.
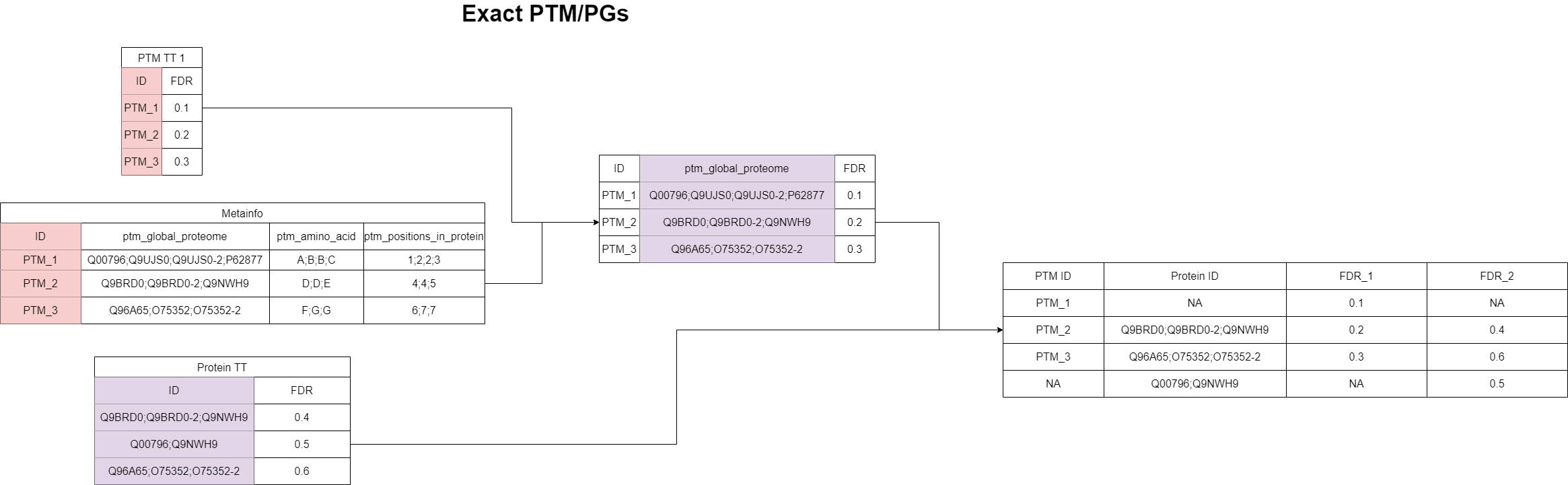
“Exact matching” for the PTM/PG case
Here, we are dealing with PTM data on the one hand and proteomics data on the the other. As before, the intention of this “exact matching” is having a high confidence when finding a match. This is not given for two unrelated peptidomics/proteomics studies, because the features were potentially identified under completely different circumstances. Hence, for doing this matching, it is required that the studies are indeed related to each other. This information can be stored in the sample table during data integration.
If the studies are related, PanHunter doesn’t use the protein groups that were identified by DiaNN, Spectronaut or Maxquant as PTM associated protein groups but instead each PTM feature is mapped using an proprietary mapping algorithm to a protein group in the proteomics dataset during data integration. These specifically inferred protein groups will then be used for mapping to the related proteomics dataset, and again, the protein groups must be fully identical to be considered a match. This also means that no isoform removal or any other modification on the protein groups are made in this process.
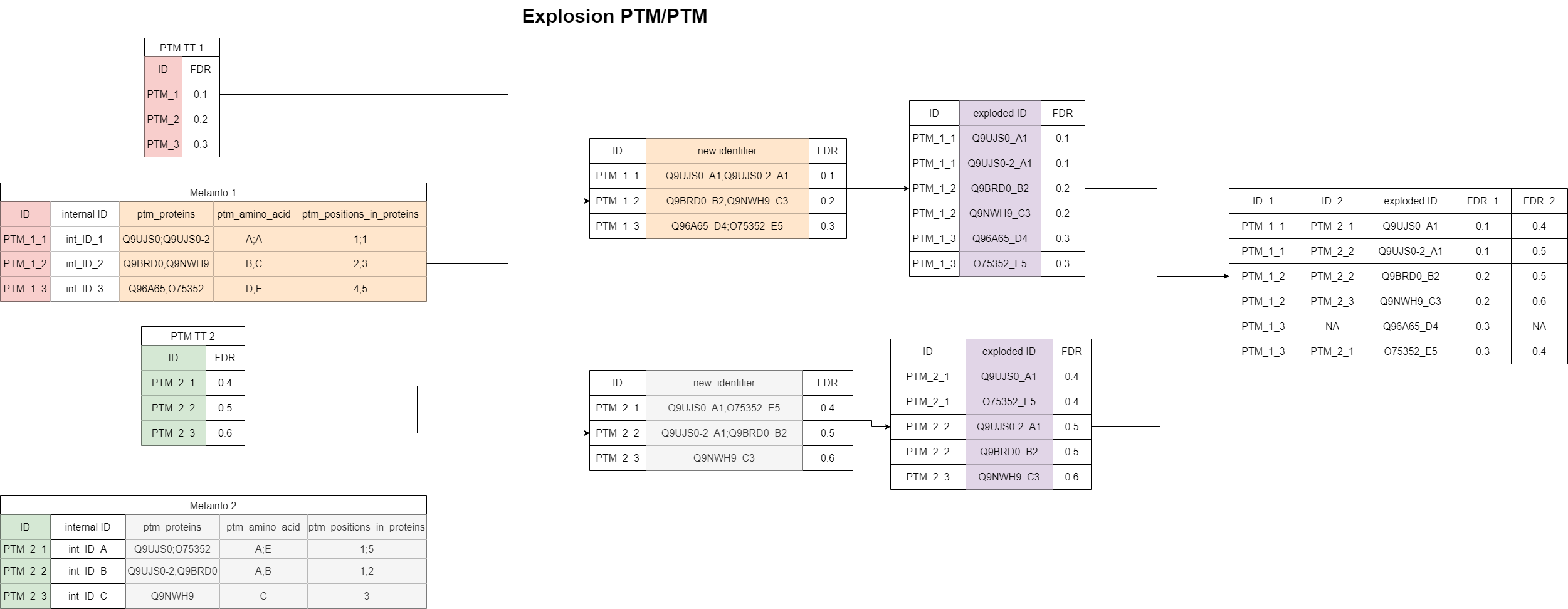
“Explosion matching” for the PTM/PTM case
Compared to the “Exact matching”, the “Explosion matching” is less strict and more matches will be found. In this context, “explosion” means that the list of associated proteins gets split-up to single proteins. Each protein now gets appended by the modified amino acid and the location of the modification. Furthermore, in case of multiple modifications, the protein is appended by each modified amino acid and location separately (e.g., Q9UPW0_K_K_40_41 results in Q9UPW0_K_40 and Q9UPW0_K_41). Two PTM features are now considered as a match, if any of these protein features are identical between them. If the overlap is larger than just one feature, the list of matched features is made unique, because the information would be redundant. This will be explained in the apps in more details. This algorithm can result in multimappers, if a PTM is part of multiple protein groups. Within this algorithm, isoforms are cleaned, such that different isoforms are considered as a match.
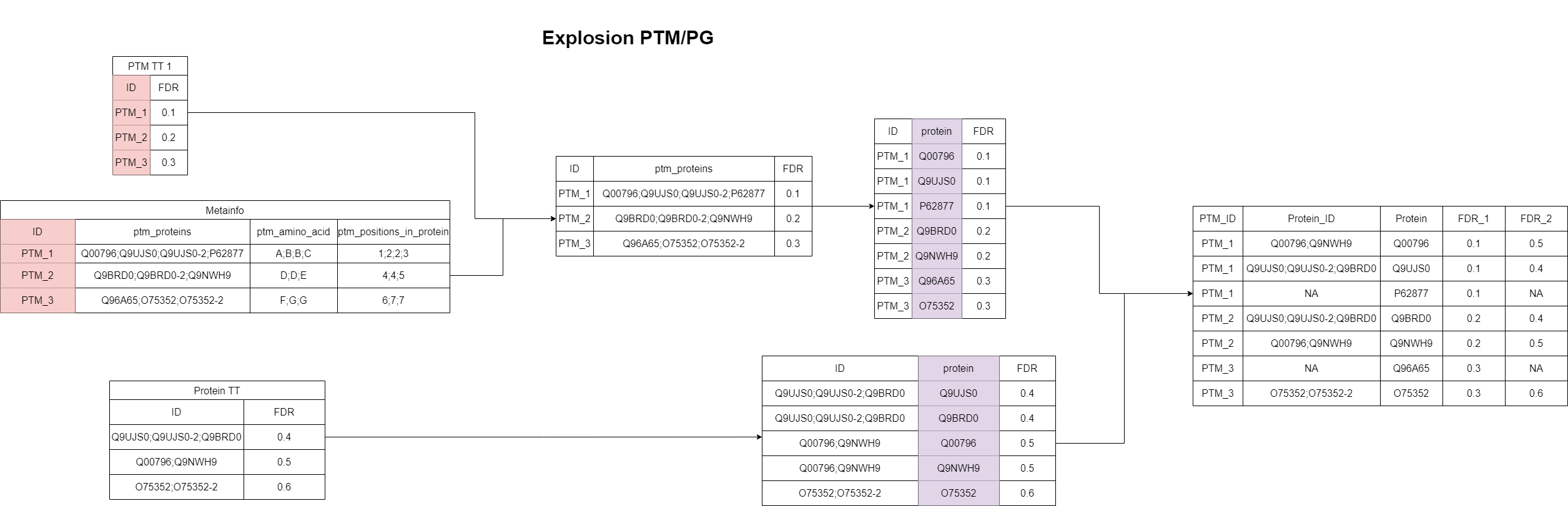
“Explosion matching” for the PTM/PG case
Since the “Explosion” algorithms are less strict, we don’t require any relations between the datasets as for the “Exact matching” algorithm for the PTM/PG case. Here again, the associated protein groups for the peptidomics study and the protein groups of the proteomics study are both split up (“exploded”). Differently than for the PTM/PTM case, here isoforms are converted to the canonical forms for both datasets. A PTM feature matches to a proteomics feature, if any of the “exploded” proteins is found in the other dataset. Again, this can result in multimappers as all PTMs of a canonical protein will match to all protein groups containing this protein.
“Explosion matching” for the PG/PG case
For the matching between two proteomics studies, always the “Explosion matching” is used. Again, for both datasets the protein groups get “exploded”. Two features match, if any single protein is found in both protein groups. Isoforms are converted to the canonical form here as well.