Data Overview
The Data Overview app helps in exploring and managing all project-related data. It provides the following tabs:
Studies tab

This tab provides a complete data overview of all the studies integrated in the project. You can use the to apply filters to the table columns to find your studies of interest.
Samples tab
In this tab, you can explore the samples available in this project. First, you need to select a study/studies in the Sample selector on the left side panel. Subsequently, the plot and the table overview will show all samples from the selected studies.
Plot Overview
The bar plot shows the distribution of samples across various variables. Using the “Group by” option splits samples into separate bars, grouped by the selected variable. For example: For the selected study in the sample selector, you can apply “Group by” = Genotype, and set “Color variable” as “Tissue” which then adds colors to the bars according to the samples
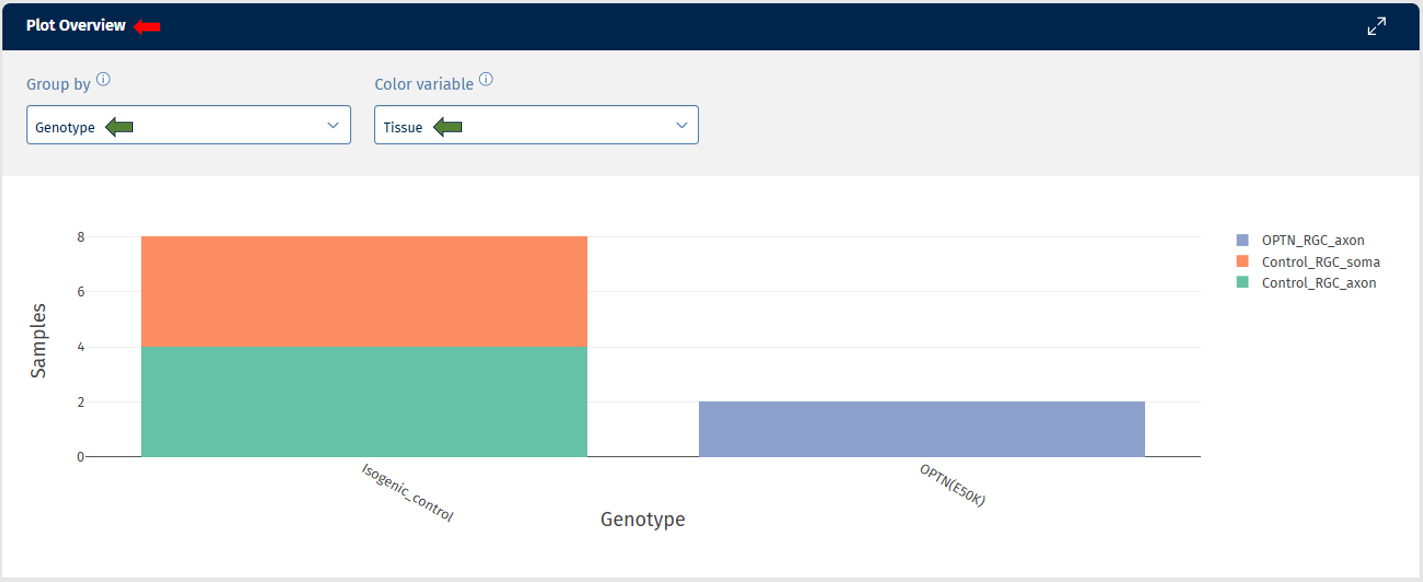
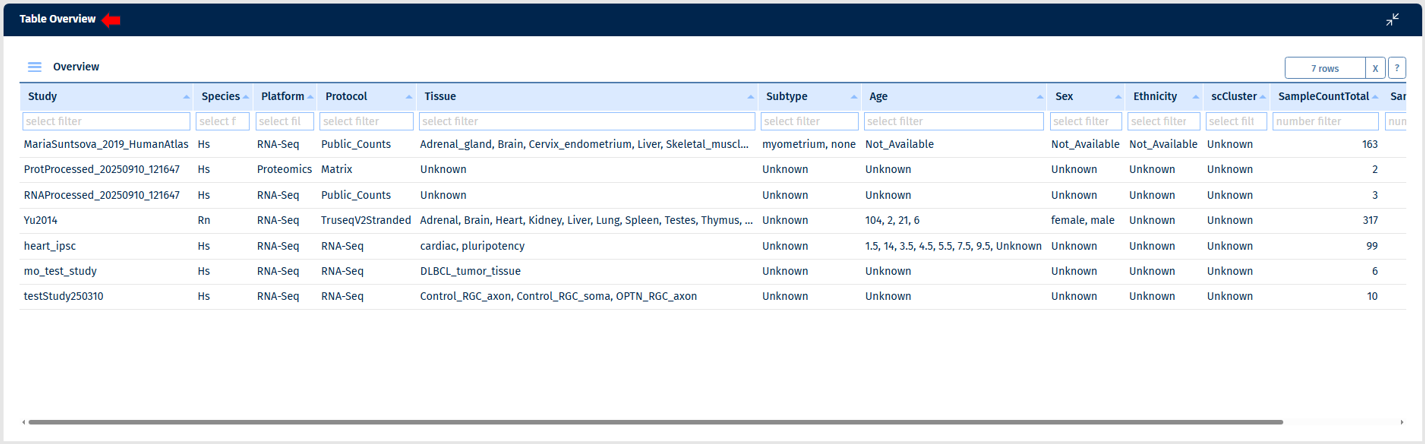
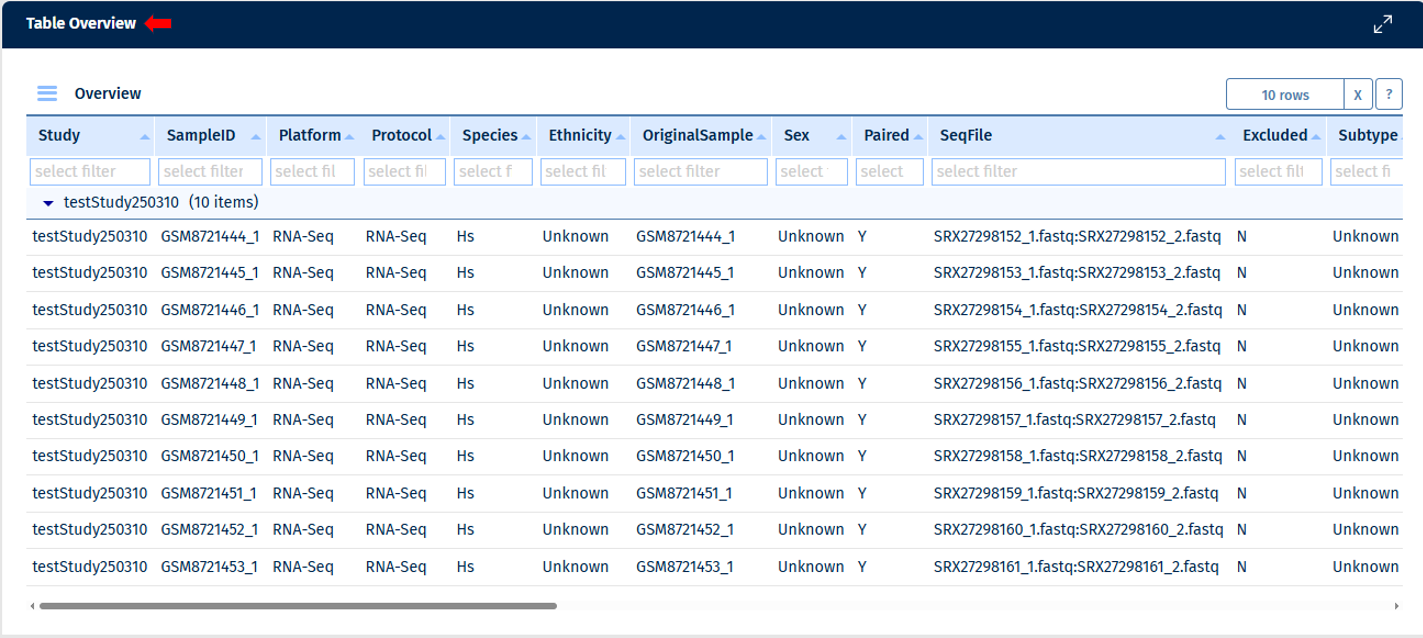
Table Overview
The samples can be analysed in a table format here, providing detailed information. Note: empty columns are not displayed in the table.
Comparisons

This tab shows an overview of all Comparisons present in the project. Here you can find information on who calculated them, when, as well as what data and parameters were used. Via the buttons on the left side panel, which are active once you selected a Comparison by clicking on a row in the table, you can directly jump into downstream analysis.
Manage Selection
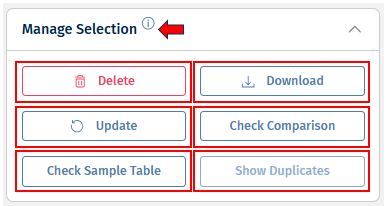
This panel contains the following buttons:
-
Delete - This option deletes a Comparison.
-
Update - This button triggers Comparison re-calculation. This is useful, for example, when new samples were added to the same study and now you want to update your Comparison.
-
Check Sample Table - This option allows users to compare sample table used during comparison computation and the sample table recreated based on the Comparison recipe. Main reason of difference between stored and recreated sample tables are updates of sample tables (e.g., addition of new samples or changing sample annotations). If differences between stored and recreated sample tables are identified, please consider updating the Comparison.
-
Check Comparison - This button initiates the check whether the Comparison saved and a Comparison recomputed based on the Comparison recipe and previously saved sample table are different (i.e. recreation of sample table is not performed as a part of this comparison). A standard reason of difference between stored and recomputed Comparisons are updates in methods for Comparison computation. If differences between stored and recomputed Comparisons are identified and these differences were not expected, please consult with the PanHunter team, or assigned bioinformaticians.
-
Show duplicates - This option searches for duplicates of the selected Comparison, which you can delete afterwards. Comparisons that have duplicates show the value “duplicated” in the “Status” column.
-
Download - This option downloads a .zip file with all Comparison data. The data is the same as in .html files opened through “Open Data” buttons.
Below is a list of files and folders found in the downloaded ZIP-file. Please note that the top-level files are always present. Folders and files in the folders vary depending on the post-processing steps that were performed while creating the Comparison.
-
DifferentialFeatureAbundance.csv : This file holds the main results of the Comparison calculation.
For each feature in the Comparison, the following values are listed, they depend on the underlying type of data (transcriptomics, proteomics, genomics, metabolomics, etc).
-
FeatureID: PanHunter feature ID.
-
EnsemblID: gene ENSEMBL ID.
-
Symbol: Gene symbol.
-
Name: Human readable name of the feature.
-
Abundance: Average abundance of the feature across denominator samples.
-
FDR: P-value adjusted for multiple testing.
-
p-value: p-value as it is reported by limma or DeSeq2.
-
SE: (optional) Standard error as it is reported by limma.
-
logFC: Log2 fold change as it is reported by limma or DeSeq2. Please note, that currently fold change shrinkage is not applied.
-
sig: Binary value, telling whether a feature is significantly regulated.
-
-
Metadata.json : This file contains Comparison metadata in JSON format. This contains the internal Comparison ID, computation date, user-id, the method and input parameters used to calculate the Comparison, list of samples used, or filter steps applied to sample table. In principle, this is the same information as displayed in the table “Comparisons Overview” in the Comparisons app.
-
Recipe.json : This file holds instructions for PanHunter about how to create the Comparison. This is a JSON formatted version of the input settings provided by the user in the New Comparison tab present in the “New Comparison app”. It includes:
-
rules for filtering the sample table
-
parameters for and type of comparison algorithm
-
post-processing steps to be carried out
-
-
SampleTable.csv : This file contains the table of samples used for calculating the Comparison. For each sample the file holds a number of properties, e.g., SampleID, Study, Platform, Protocol, Species. Other properties are dependent on the underlying experiment and type of sample.
-
Enrichments : This folder contains the results of the “GO enrichment” and “Pathway enrichment” post-processing steps.
- Gene Ontology
The information in these files describes the results of enrichment analyses for the GO gene sets based on the features found to be significantly regulated in the Comparison.
For each domain of the GO one file is provided:
-
GOBP.csv - GO terms for biological processes
-
GOCC.csv - GO terms for cellular component
-
GOMF.csv - GO terms for molecular function
Please find more information about Gene Ontology (GO) database in Enrichment Visualization app.
- Wikipathways
The information in these files describes the results of enrichment analyses for the Wikipathways gene sets based on the features found to be significantly regulated in the Comparison. For example, it provides statistical values from the Wilcoxon, Kolmogorov-Smirnov and Fisher (exact) tests and was computed based on data from Wikipathways. For each available species, a separate file is provided.
For example:
- Wikipathways_Rn.csv - This file provides information about organism-specific pathways for Rattus norvegicus
Please see Pathway Visualization App documentation for more information.
-
FilteredOut : This folder contains CSV file for the features filtered out based on their abundance across the samples used in the Comparison.
For example:
-
ModelBased.csv - This file contains all features that were removed by the model-based filtration step.
-
Networks : This folder contains the results of the “Subnetwork extraction” post-processing step.
-
Biogrid : The files in this folder holds information about the gene/protein interaction networks enriched with the features found to be significantly regulated in the Comparison.
For each available species, a file is provided with references to subnetworks in the Biological General Repository for Interaction Datasets (BioGRID).
For example:
-
Hs.csv - Homo sapiens
-
Rn.csv - Rattus norvegicus
Please see Network Visualization App) documentation for more information.
- Signatures : This folder contains the result of the “Signature analysis” post-processing step. There is an overview file with summary information and one file for each signature collection analysis that has been carried out. The latter contain various statistics and tests in order to identify signatures that are similar or opposite to the Comparison results.
For example:
-
Overview.csv - This Overview file consists of the overview data with the signature collections for which the analyses was done.
-
ManualSingleDrugPerturbations.csv - This file consists of the results of signature analyses for a particular signature collection (“ManualSingleDrugPerturbations” in this case). Each row in this file represents an individual signature, its annotation, and results of the directed enrichment analyses based on the features found to be significantly regulated in the Comparison.
Please see [Signature Visualization App]((/apps/standard_apps/signatures/) documentation for more information.
- TFTargets : This folder contains the results of the “TF analysis” post-processing step. The files contain several statistical values to identify Transcription Factors (TFs) whose target genes are overrepresented in the Comparison.
For example:
ChipAtlas.csv - This data is compiled by utilizing the ChipAtlas dataset.
Open in APP
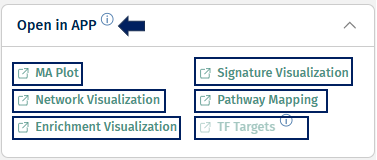
This panel provides quick links to other PanHunter apps where the selected comparison can be further explored. Please note that selection of available apps may vary, depending on data available for the comparison.
Open Data
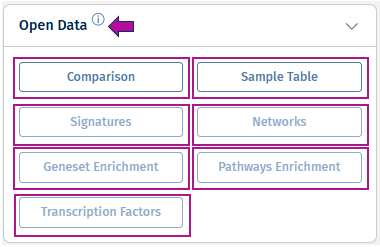
This panel shows what data is available for download for the selected Comparison. If some post-processing steps were skipped during a Comparison calculation, the corresponding data is then not available and the buttons are greyed out. The full list of data includes the following:
-
Comparison - This option opens the results of differential abundance analyses (fold changes, p-value, FDR) for the features found significantly regulated.
-
Sample Table - This option shows an overview of the samples used for the Comparison calculation.
-
Geneset Enrichment - This option displays the results of gene ontology term enrichment analysis.
-
Pathway Enrichment - This option provides an overview of pathways of which the differentially abundant features are members. Calculated based on the gage and GeneAnswers R-packages.
-
Signatures - This option displays additional information on which known compounds leads to similar differentially abundant features.
-
Transcription Factors - This option shows downstream analysis of the transcription factors that might be relevant to the changes in feature abundances observed in this Comparison.
-
Networks - This option shows the gene interaction networks enriched with differentially regulated features.
Comparison Groups

This tab shows an overview of Comparison Groups, which is a set of Comparisons that were generated using very similar parameters. Currently, you can only generate a Comparison Group using RStudio or by requesting it from BIX colleagues.
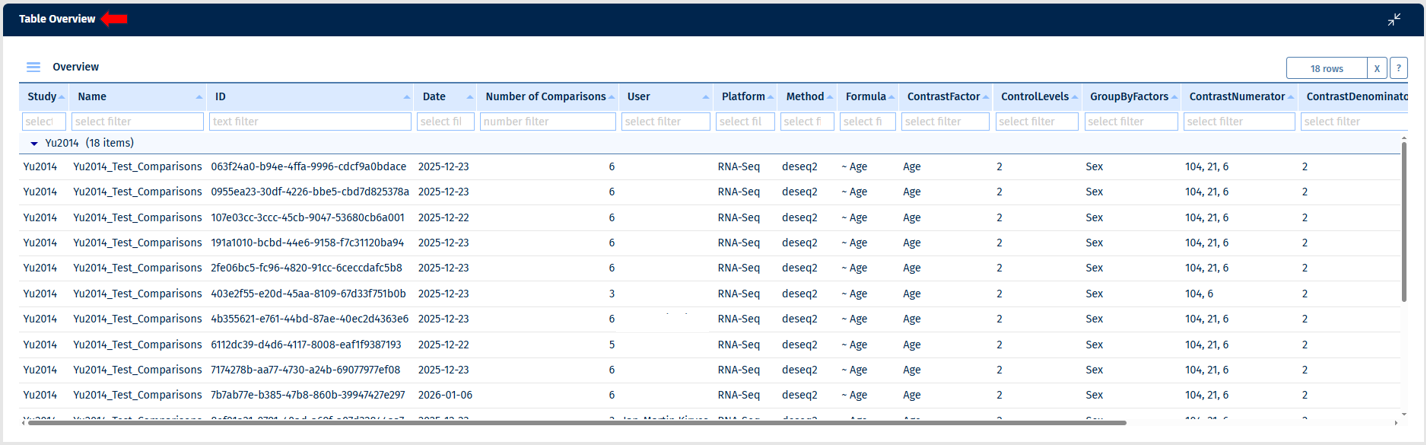
In the overview, you can see who created a Comparison Group, when, how many Comparisons it contains, and what parameters were used for its calculation.
Feature Lists
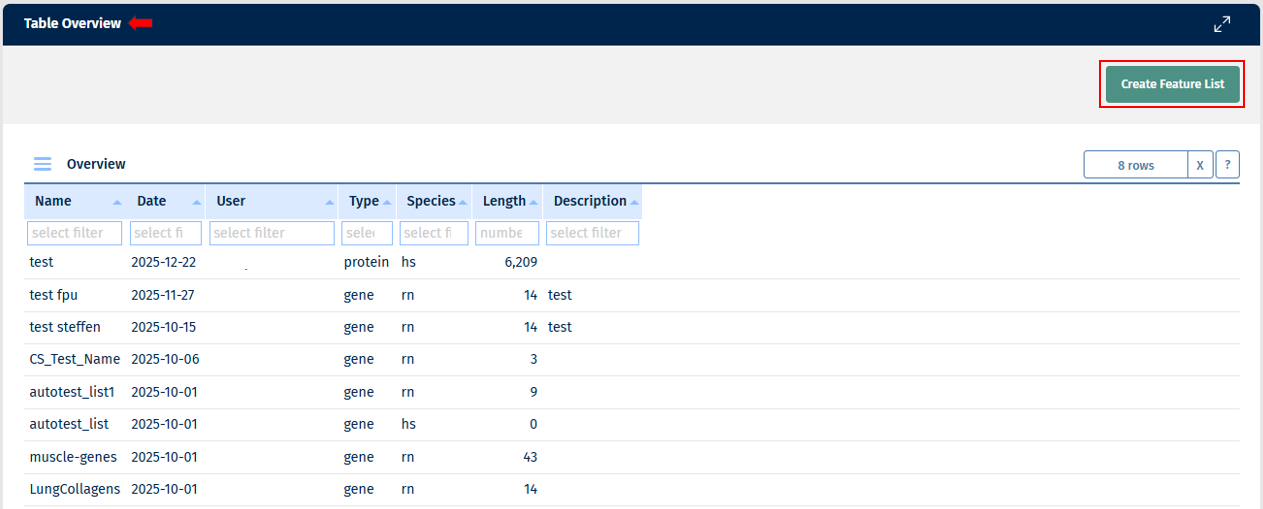
This tab shows an overview of Feature Lists available in this project. Feature Lists are just a list of Feature IDs. Here you can see who created it, when, what type of Feature List (gene, protein, etc.), what species, how many entries the Feature List has, and it’s description. Whenever you need to select features in an app, there is an option to provide a Feature List as an input.
There is also a possibility to create a new Feature List in this app. By clicking the “Create Feature List” button, you will see this pop-up:
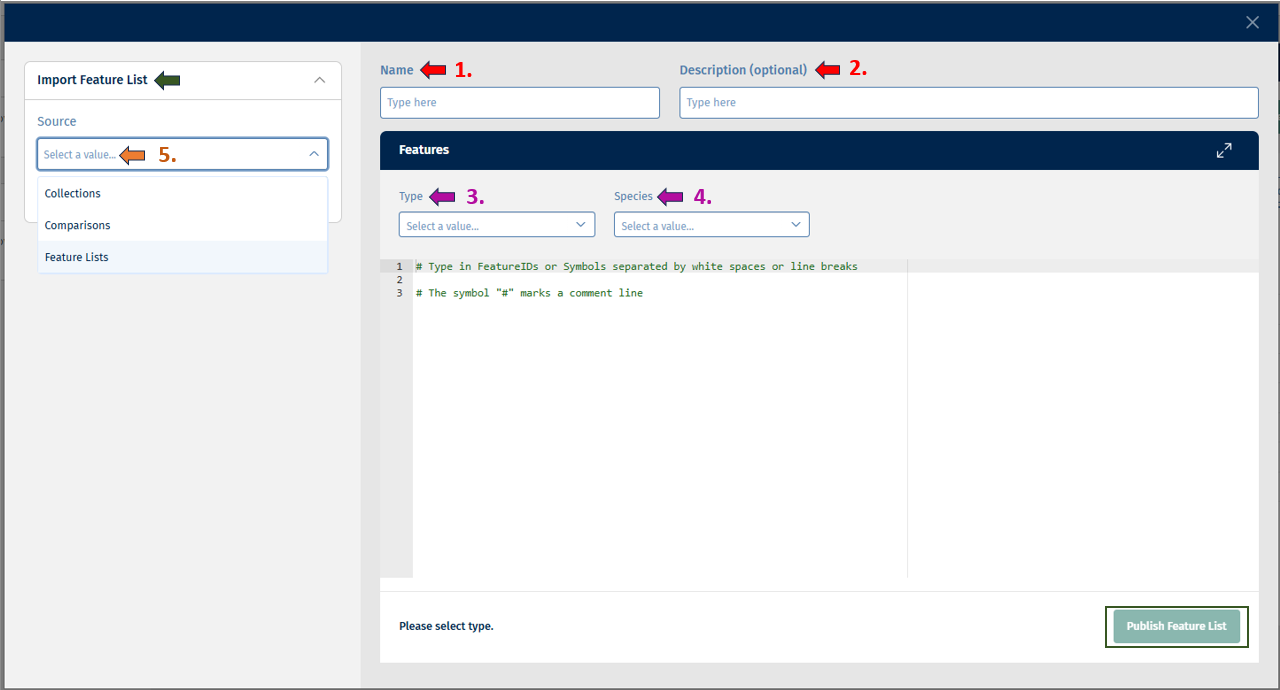
Before creating or saving a Feature List, you must enter a name and optionally a description for your list.
You must provide the “Type” which includes (Gene, Protein, etc.) and “Species” such as (Hs, Mm, Rn) information. Following this, you can access the below-given sources from the Import Feature List panel for creating your feature list:
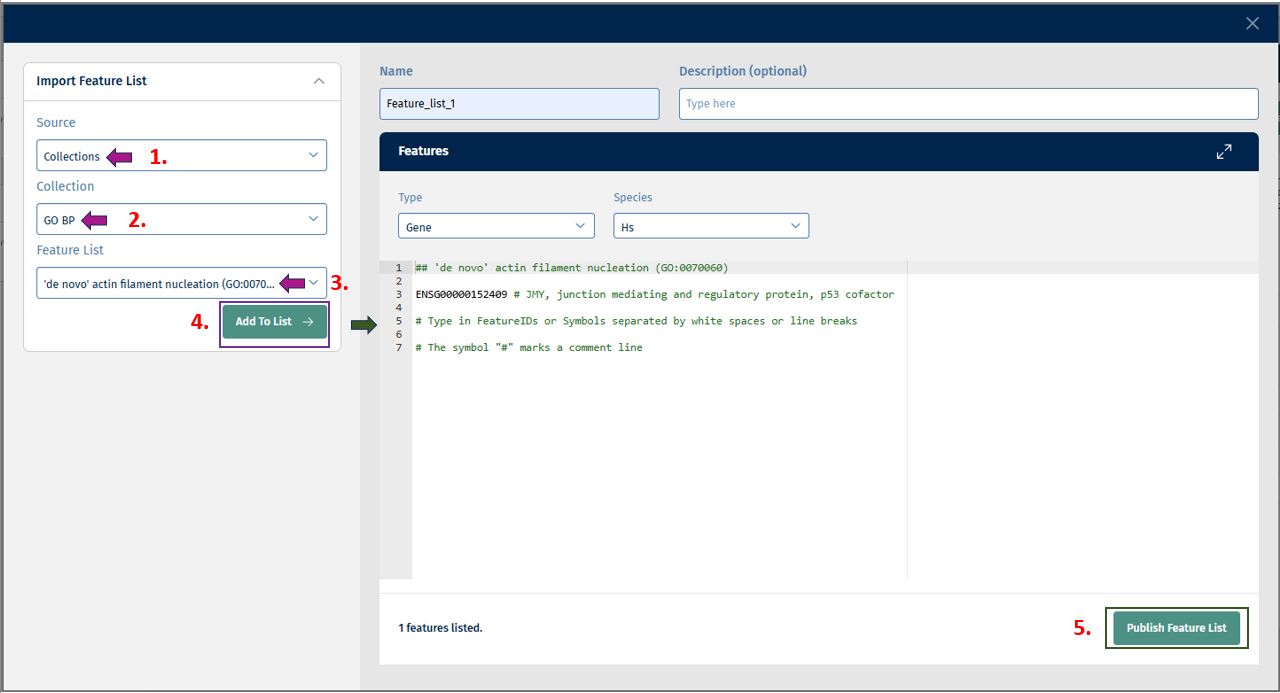
- Collections: This source displays a list of collections such as GO BP, GO CC, GO MF, etc to choose from. After selecting from this list, you get the option of “Feature List” from which you can select the feature/s of interest and add it to the list.
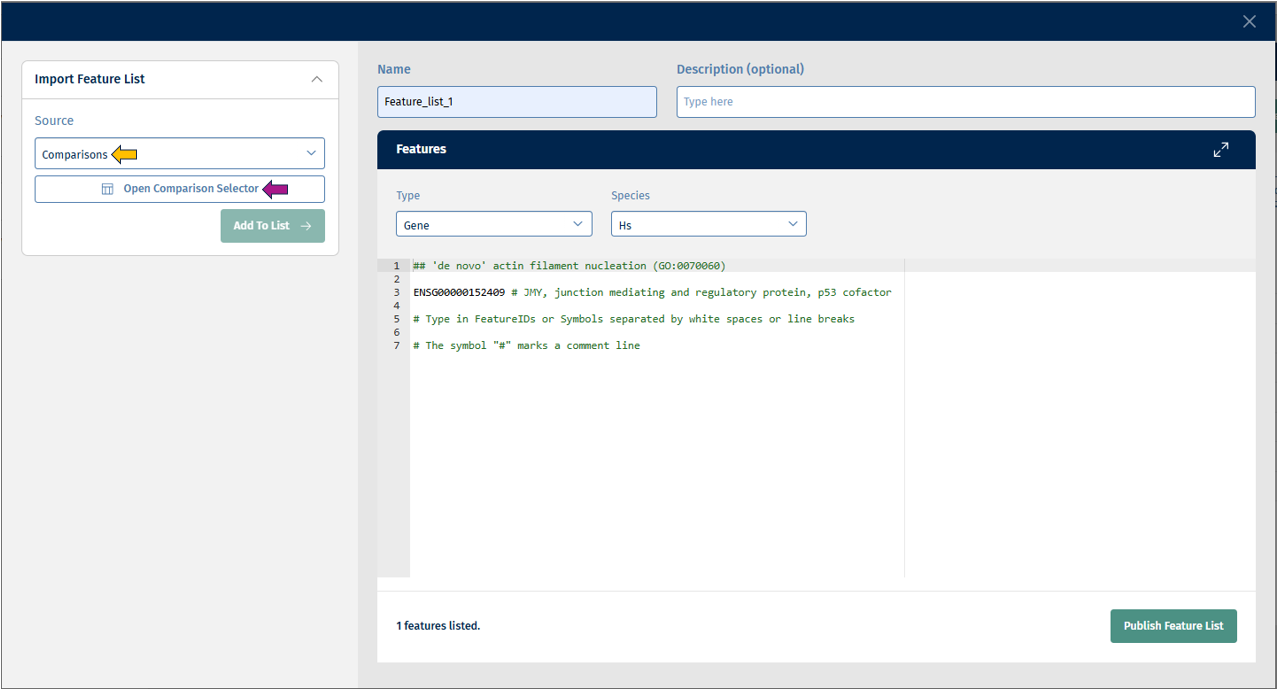
- Comparisons: This source provides you with a comparison selector called “Open Comparison Selector”.
Clicking on this, opens a view with options “Single Comparisons” or “Comparisons from Groups” to choose from. You can choose your comparison study/-ies of interest and click on “Confirm” to add it to your Feature List.
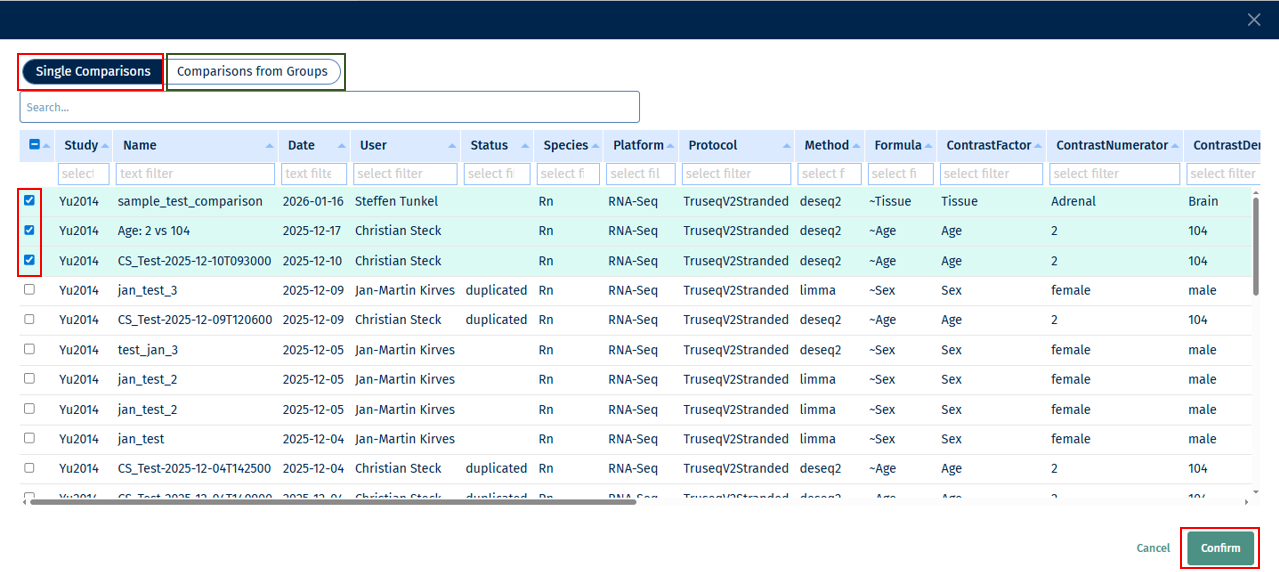
Note: You can choose study/-ies from only one of the options (“Single Comparisons” or “Comparisons from Groups”) at a time.
- Feature Lists: This source provides you with a feature list selector called Open Feature List Selector from which you can select your Feature/s of interest and add it to the list.
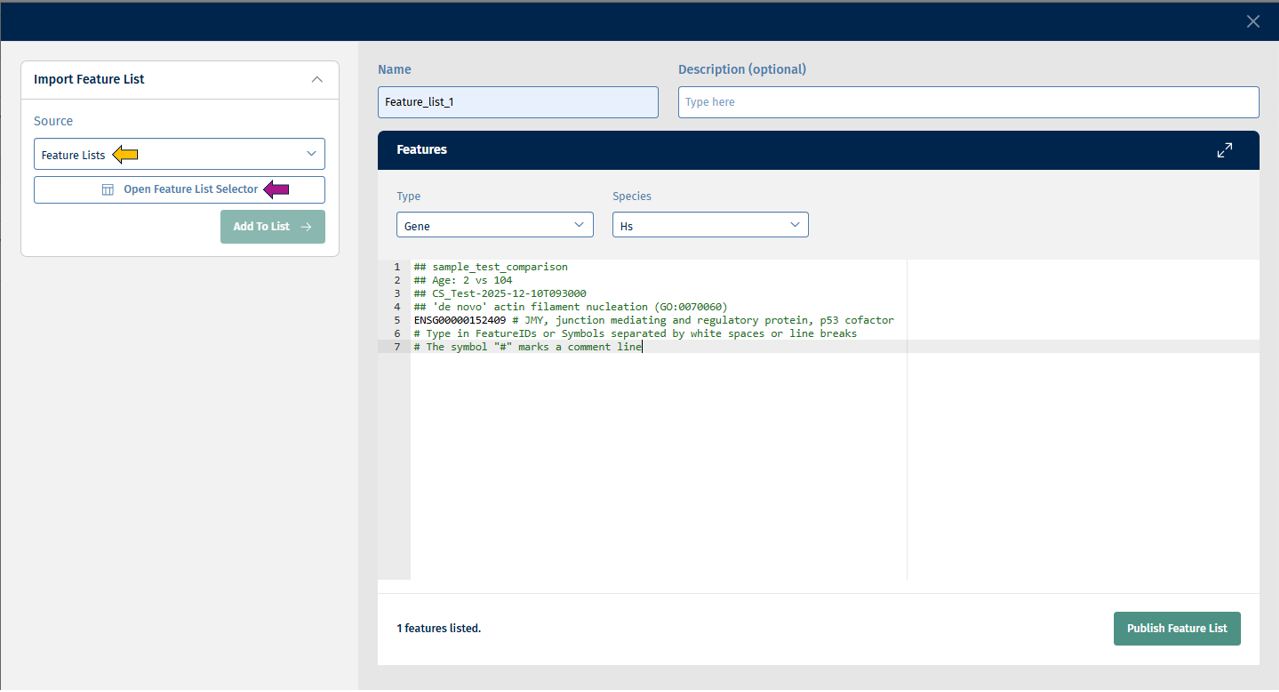
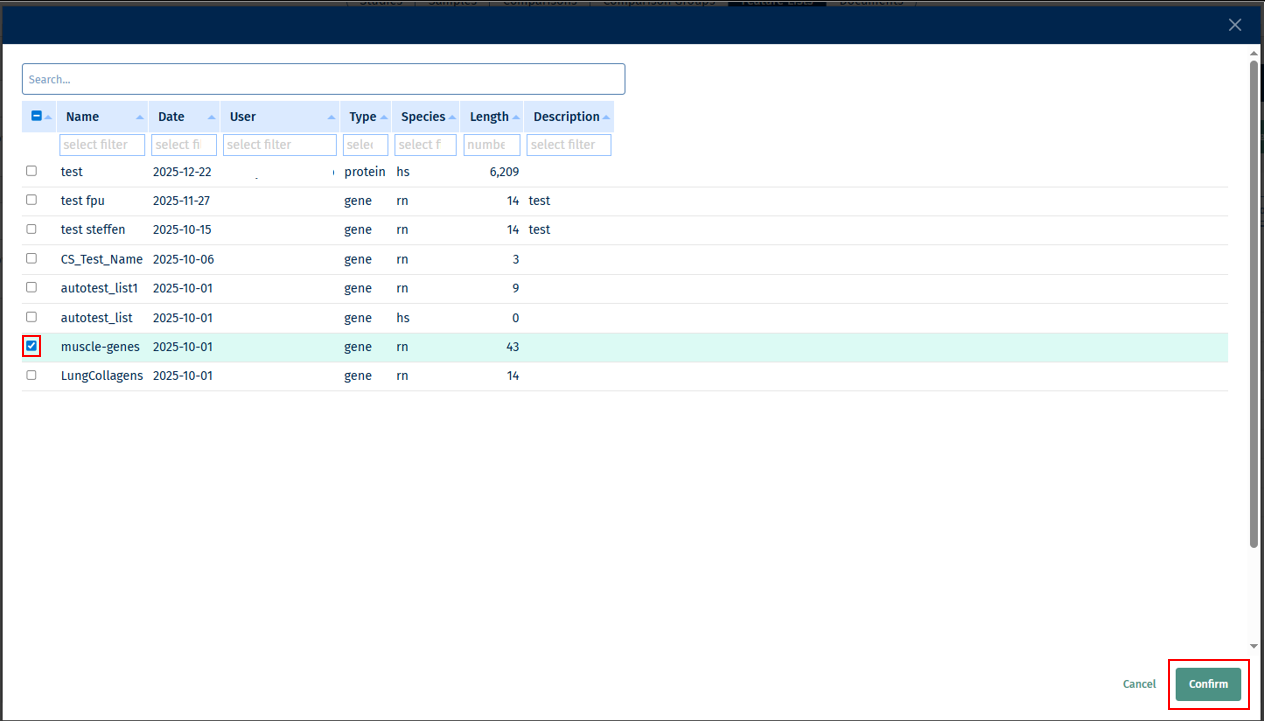
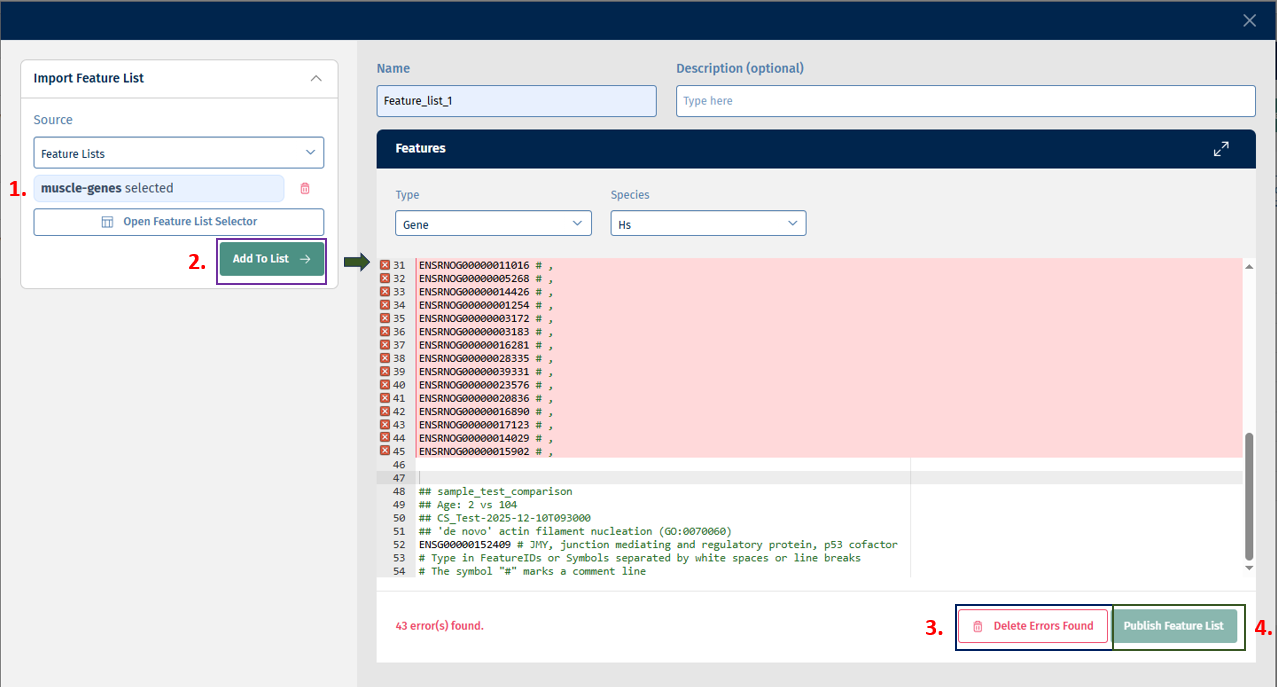
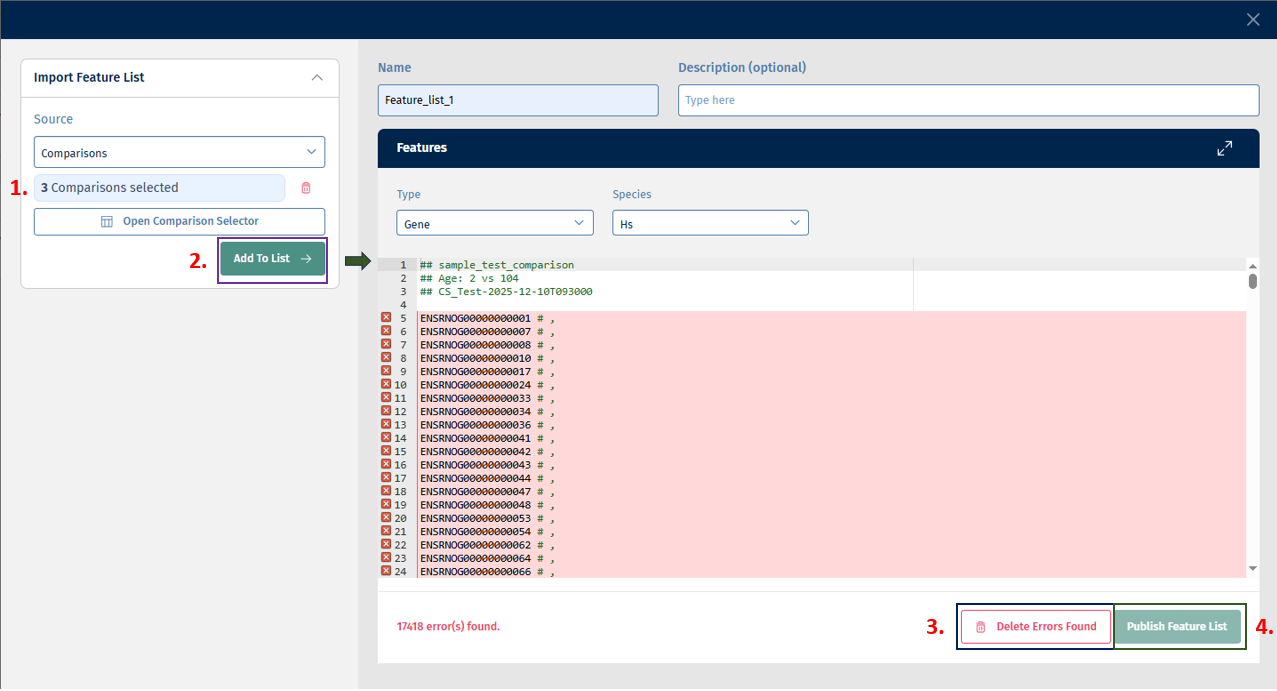
Click on “Add to List” button to add the above selected features to the list. Finally, click on the Publish Feature List button to export your curated feature list.
You can also combine multiple sources in one Feature List. Alternatively, you can also type Feature IDs in the text field.
There is an error correction mechanism that shows errors when Feature IDs are not mappable to the selected type or species: You can then either correct the errors or delete all error-prone entries using the Delete Errors Found option.
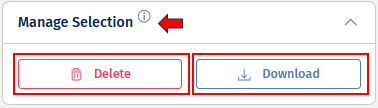
With the help of Manage Selection panel on the left side, you can delete, and download a Feature List, and the Open Data panel, helps to open a Feature List.
Documents

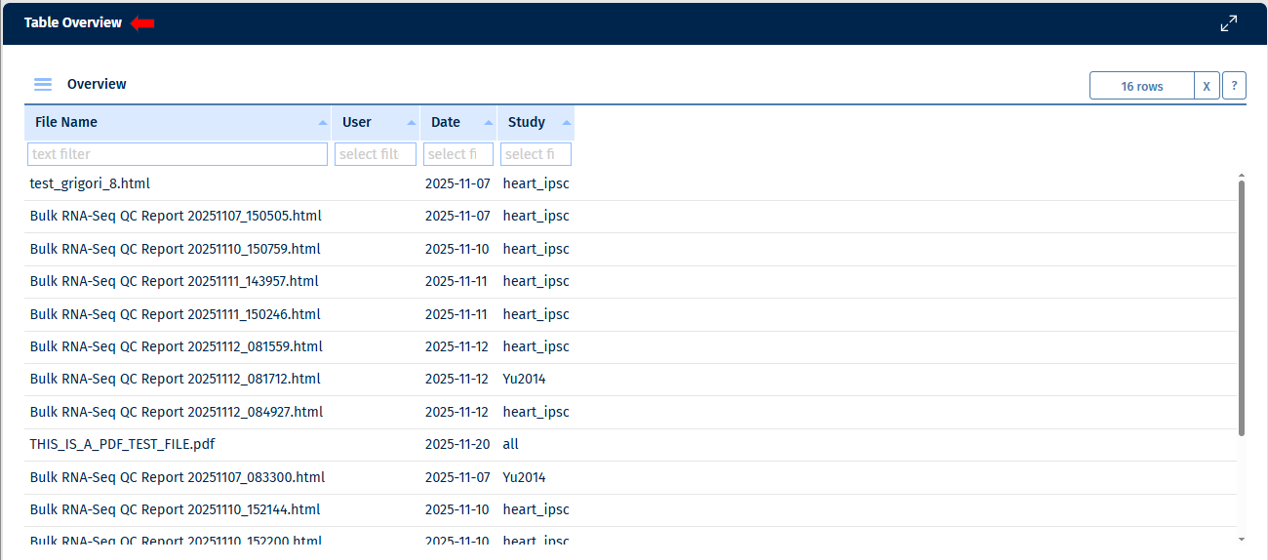
This tab shows an overview of documents available for this project. Here you can often find automatically added .html reports as well as manually uploaded documents (.txt, .pdf, MS office formats, etc).
You can upload a document using the Upload File panel where you need to specify if it is study or project-related. If you choose study-related, multiple projects with the selected study will see this document. If you choose project-related, this document won’t be shared with other projects.
Additionally, using the Manage selection panel you can delete, download, or open files.