This section explains to the users how to perform the following data management activities:
- Overview of available studies and their upload status
- Uploading, updating and deleting data
This the multi-page printable view of this section. Click here to print.
This section explains to the users how to perform the following data management activities:
For the following species reference genomes are available in PanHunter: TO-DO give list
For model species such as mouse and human we maintain several reference genomes. If you would like to know which ensemble version is the latest in the respective release please see our changelog. The reference genome consists of a fasta file that represents the actual DNA strands and an annotation file (gtf file) that contains the positional information of every gene in the genome/fasta file.
For the human reference genome the soft masked (sm) primary assembly is used and this is then extended at Evotec with custom spike-in sequences. Soft-masked means that the nucleotides for repeat stretches are converted to lower-case. Repeats can also be masked (rm), then repeat associated nucleotides are converted to N's. The primary assembly tracks a single unbranched path through the genome. In other words there is only a single base per position and the so called haplotype (alternative base calls) are not included. Toplevel fasta files also include alternative base calls/ haplotype information.
One of the spike-ins, that the soft-masked primary assembly is extended with at Evotec, is PhiX174. PhiX174 is often used in illumina sequencing runs to increase the library quality or balance the GC content (See Phix Illumina version3 - product by illumina). Please note that the reads for PhiX should not be assigned to the fastQ files because there is no index read attached to the PhiX transcripts. Sometimes these reads do get erroneously assigned to a fastQ file due to index read bleeding (the index of a closeby cluster on the flowcell is interpreted as the index of PhiX). Barcode hopping could the other reason for an erronous assignment, this happens when indices break of and reattach within the multiplexed libraries.
The fasta file and gtf annotation contain spike in information for PhiX and for EGFP as well as for 92 ERCC (External RNA Controls Consortium) spike ins that are used to control for variation in RNA sequencing experiments.
The Data tab lists all studies of the project and indicates the state of samples for each study. Initially, for the newly created project, there will no studies listed:
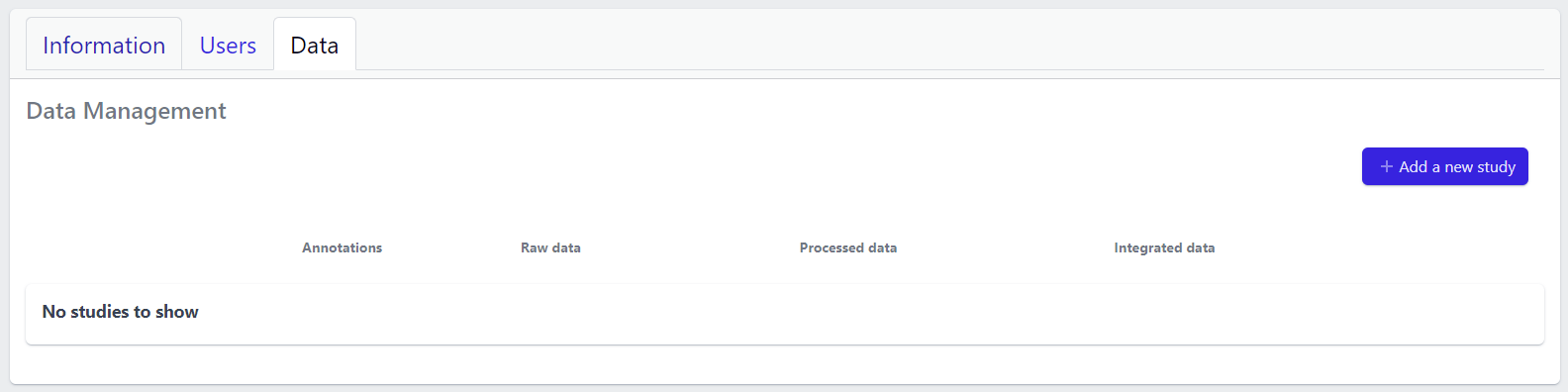
In the upper right corner an Add a new study button allows creation of new studies. Please see here for more details on how to add a new study to the project.
Once studies have been created, the page shows a list of studies:
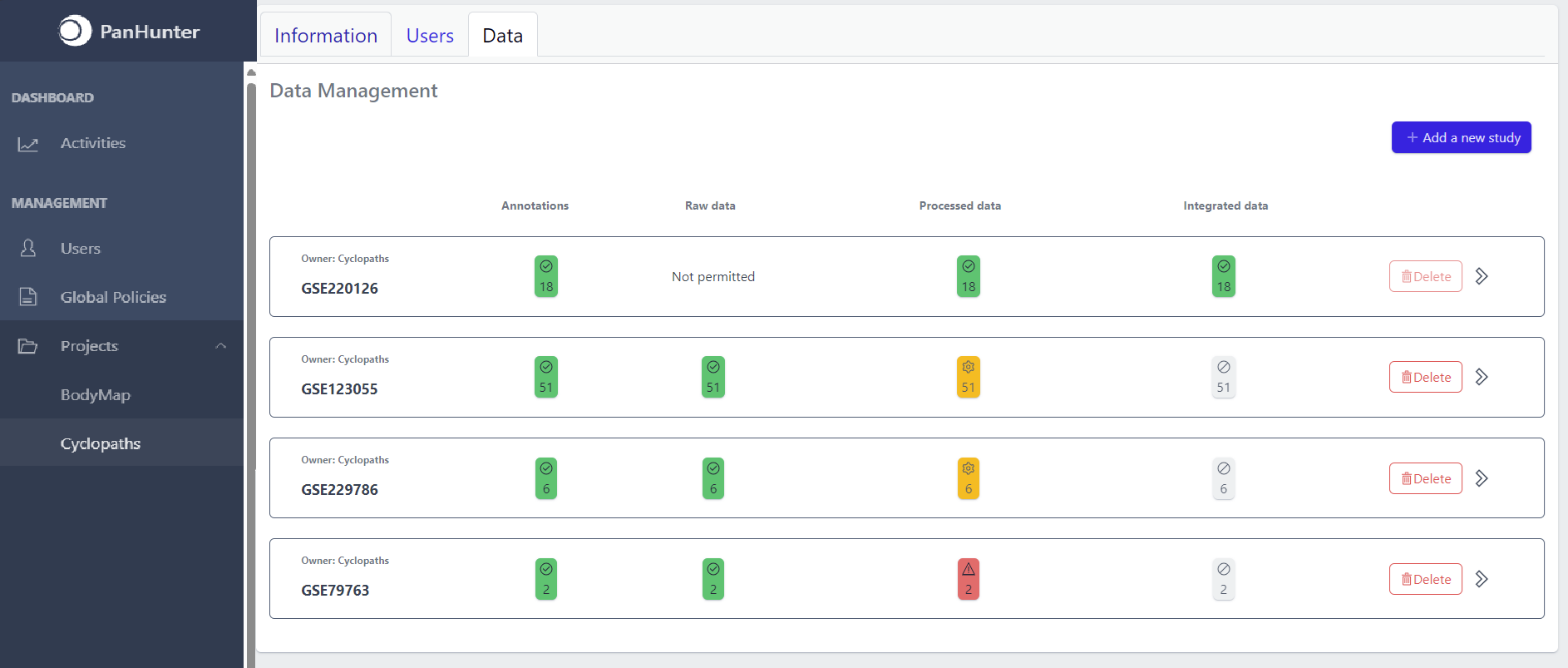
Each row corresponds to the single study, with a graphical visualization of data upload pipeline through the different stages of sample lifecycle:
The colour of the tiles indicates the status for each data upload stage described above:
Clicking on a study row leads to the detailed view of the study, which shows the same information as the overview page, but for a single study. Here, the tiles can be unfolded with a click, displaying more information about the current status of the specific data upload stage.
Additionally, the detailed view of the study enables user to perform operations like sample import, raw data inport, data processing and integration.
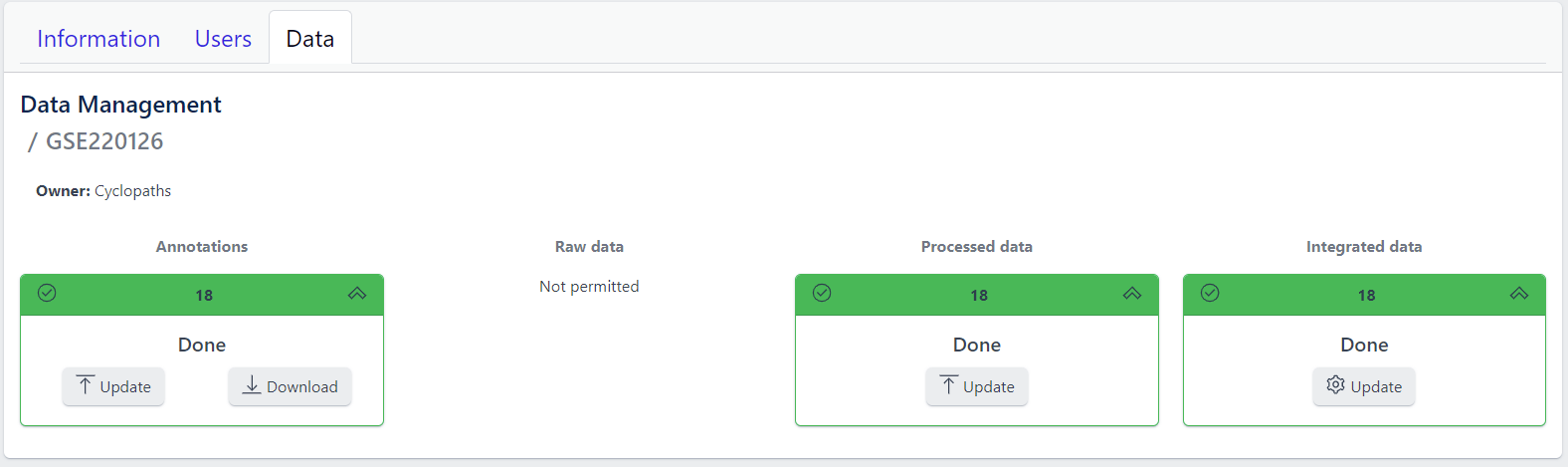
To navigate back to the overview page, click on the Data Management at the top left of the content area.
Step 1: New studies can be created in the project by clicking on the Add a new study button in the top right corner of the data overview page.
A pop-up window appears:
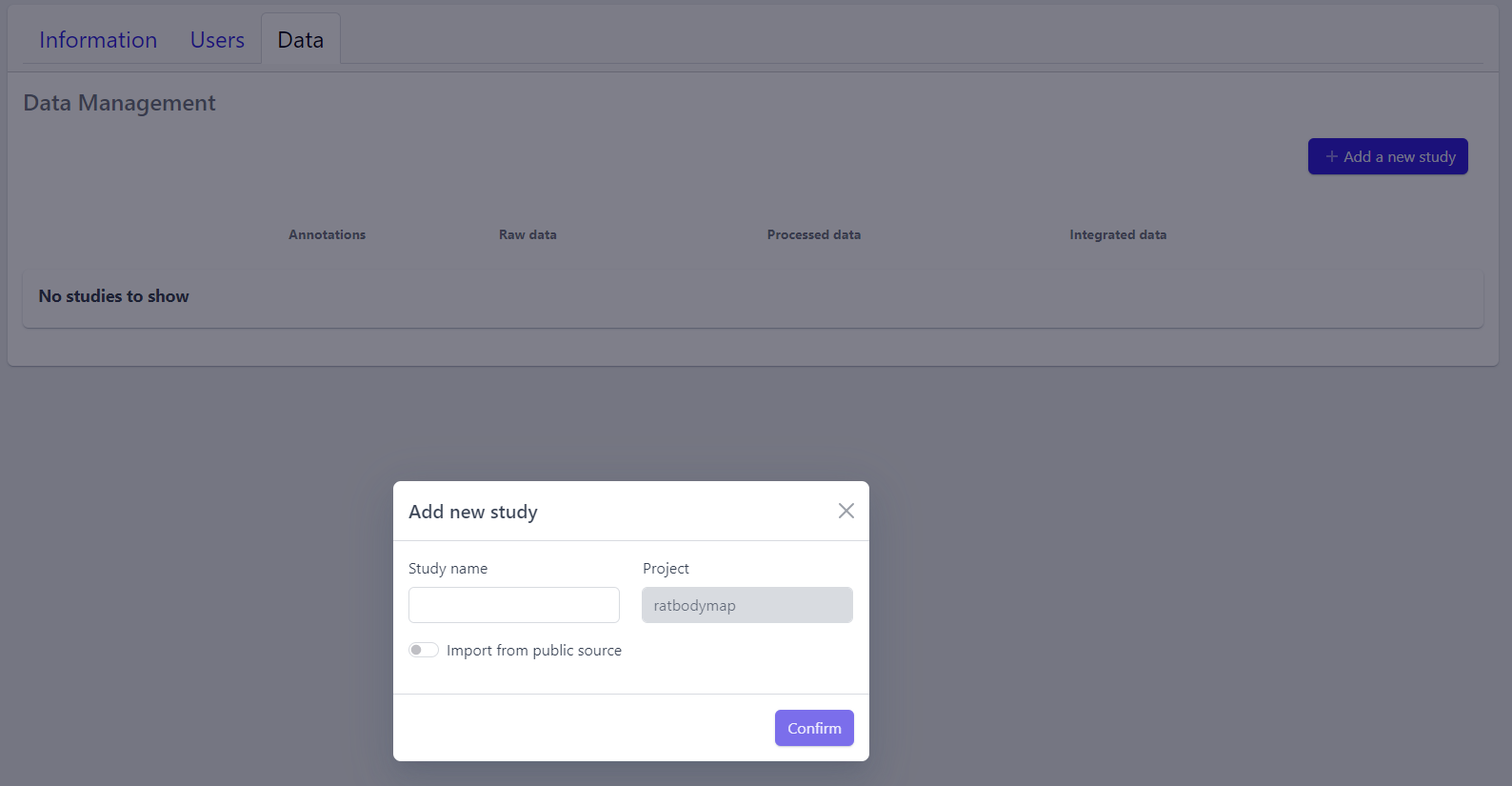
Based on the source the data is coming from, the process can be slightly different:
To upload proprietary data or other custom datasets, the process is as follows:
Step 2: Enter a study name in the pop-up window
Step 3: Click on Confirm
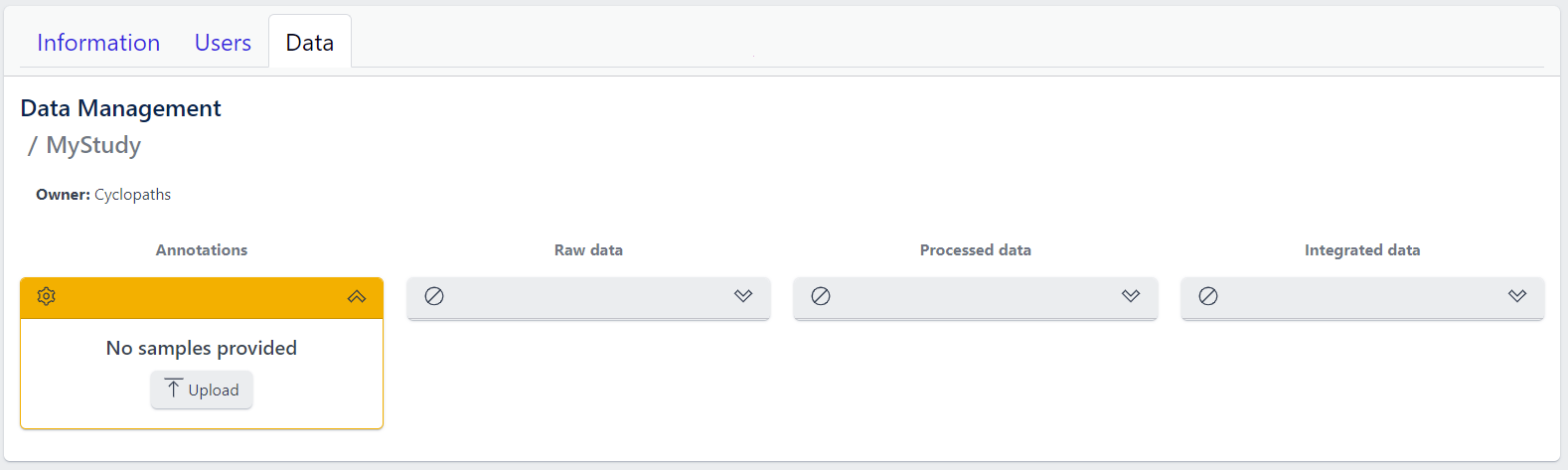
The new study will appear in the studies overview table as a new row, but without any associated samples. The Annotations tile will be coloured yellow with No samples provided status, indicating that the pipeline is ready for sample table upload.
However, with PanHunter it is also possible to import studies directly from public sources.
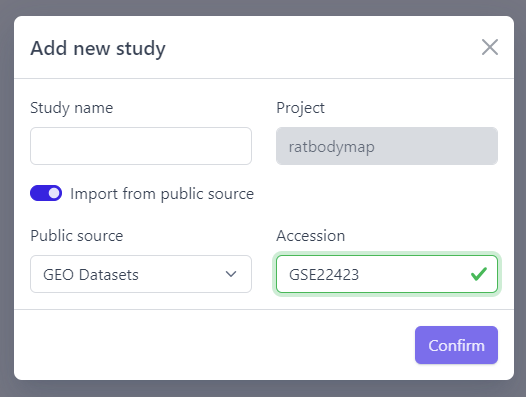
Direct import from public sources is at the moment supported for Gene Expression Omnibus (GEO), a genomics and transcriptomics database, that freely distributes high-throughput expression data submitted by the scientific community. Datasets are identified by so-called accession numbers (e.g. GSE22423).
Downloading GEO dataset directly from the database can be done with few additional steps:
Step 2: Activate Import from public source option
Step 3: Select GEO Datasets as a Public source
Step 4: Enter a valid GEO Accession number
Step 5: Click on Confirm
Panhunter will search for the given dataset accession number in GEO and download sample metadata. If successful, a pop-up window for sample table validation appears and user can proceed with Step 2. of sample table validation.
Once study is successfully created, and Annotations tile is yellow with No samples provided status. This means that we need to upload samples, which is done via sample table upload and validation.
Step 1: To upload samples from a sample table Excel file, click on the Upload button on the Annotations tile and select a file from your computer to import.
📝 In case study is imported from public sources, this step is done automatically.
Step 2: Sample table validation
After the uploaded file is successfully read by PanHunter, a pop-up window to perform sample table validation appears, as displayed below:
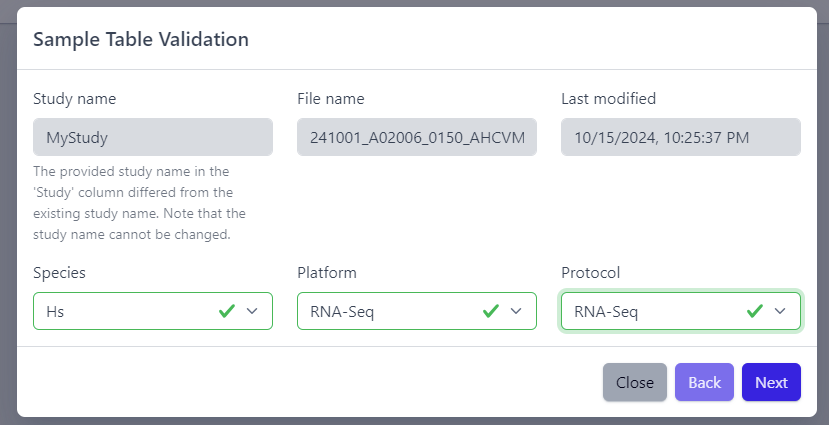
Please note that the Study name, File name and Last modified can not be edited directly. The Study name is defined during the creation of the study, and is taken from the Study column in the sample table file. File name and Last modified are defined by the file itself. To change the study name, please update the sample table file localy and restart the upload.
After you confirmed that Species, Platform and Protocol are correctly preselected, you can start the process of sample table validation by clicking on the Next button.
The result of sample table validation are shown in the same pop-up window:
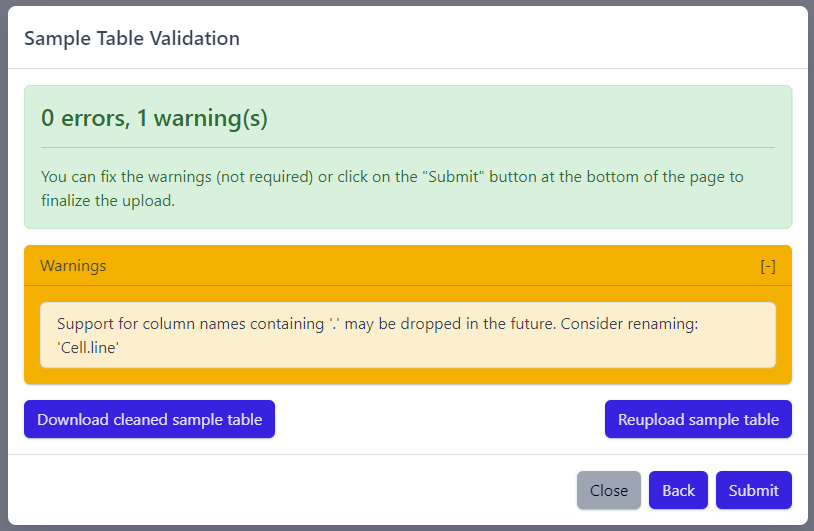
In case of errors (red), the sample table needs to be adapted. If this is the case, it is possible to use the Download cleaned sample table button to get the sample table file and modify it locally. Once sample table is modified as needed, click on the Reupload sample table button to upload the new version. The validation will be run again.
Please note that fixing warnings (yellow) is not required, but is recommended.
Step 3: Finish sample import
Once the validation passed without errors, click on the Submit button and samples will be imported and added to the study. Successfull upload of samples and study metadata will be indicated in the study details view as a green Annotations tile and the yellow Raw data tile, which indicates that the pipeline is ready for import of raw data.
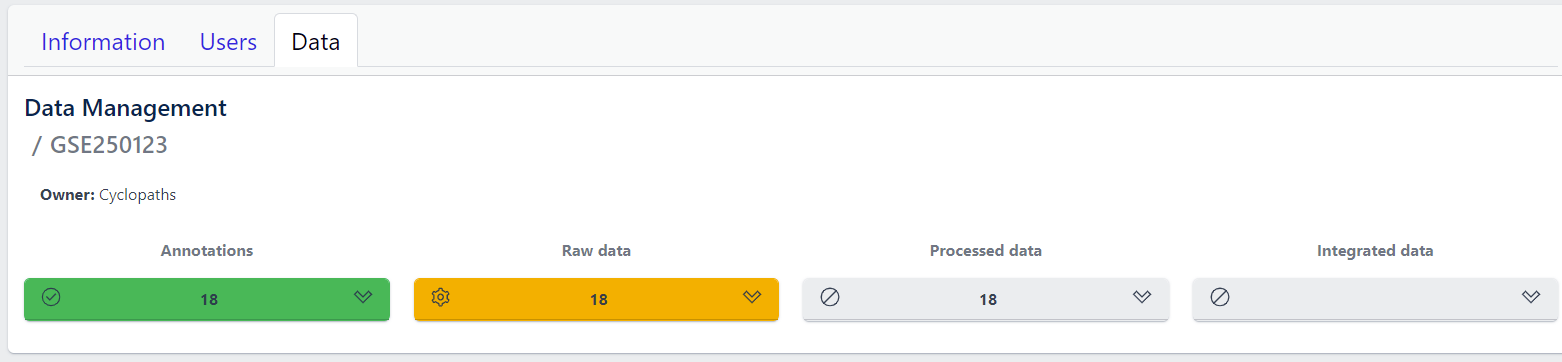
After studies are successfully created and samples are uploaded, the yellow coloured Raw data tile indicates that the pipeline is ready for upload of raw data.
📝 Info: Import of raw data via the user interface is currently supported for GEO (link) and proteomics datasets. In case you have other types of data, please contact PanHunter Support.
Step 1: Expand yellow Raw data tab in the detailed study view
Step 2: Click on the Import button
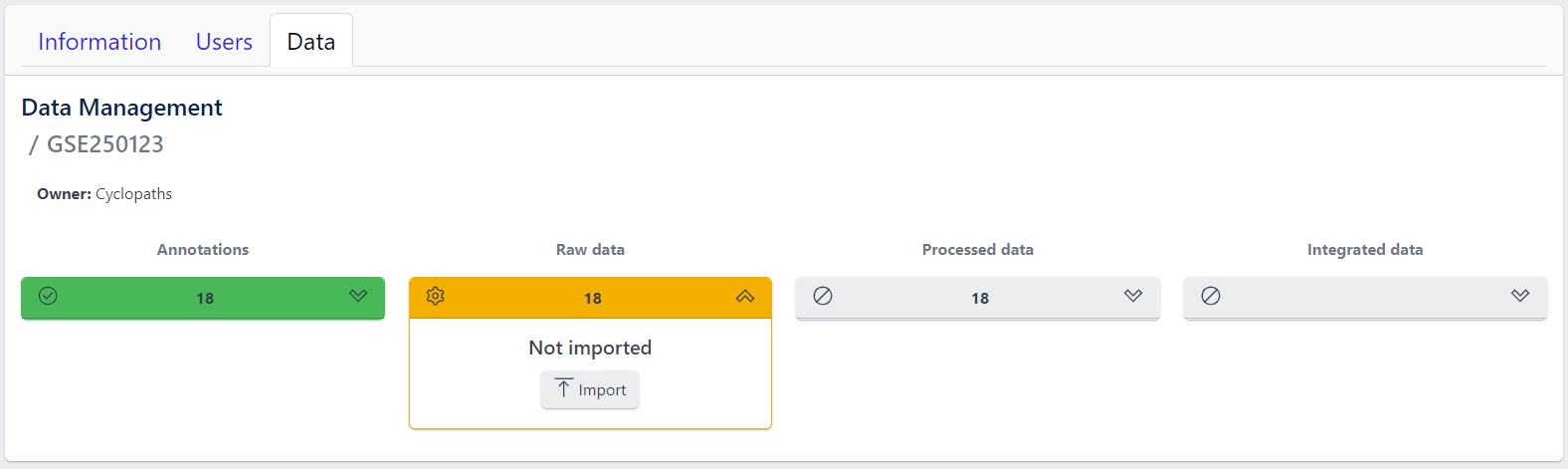
To import output files of mass spectometry instruments, please fill in all required information in the pop up window and select files from local storage to be uploaded.
💡 This section is currently in progress - for more information please contact PanHunter Support.
Once import of raw data is initiated, a background job is started on the server. Blue coloured Raw data indicates that the process is running.
Once the import of raw data is sucessfully completed, Raw data stage is coloured green and displays the number of samples for which raw data is available. Additionally, Processed data tile is coloured yellow, indicating that the data is ready for processing.
In case an error occurs, it is indicated by the red Raw data tile that can be expanded via click to investigate the job output.
Once raw data is available for imported samples, you can proceed with data processing. The data is ready for processing once Annotations and Raw data stages are successfully finished, and thus coloured green, and Processed data is coloured yellow with Not processed status.
Step 1: Unfold Processed data tile in the detailed study view.
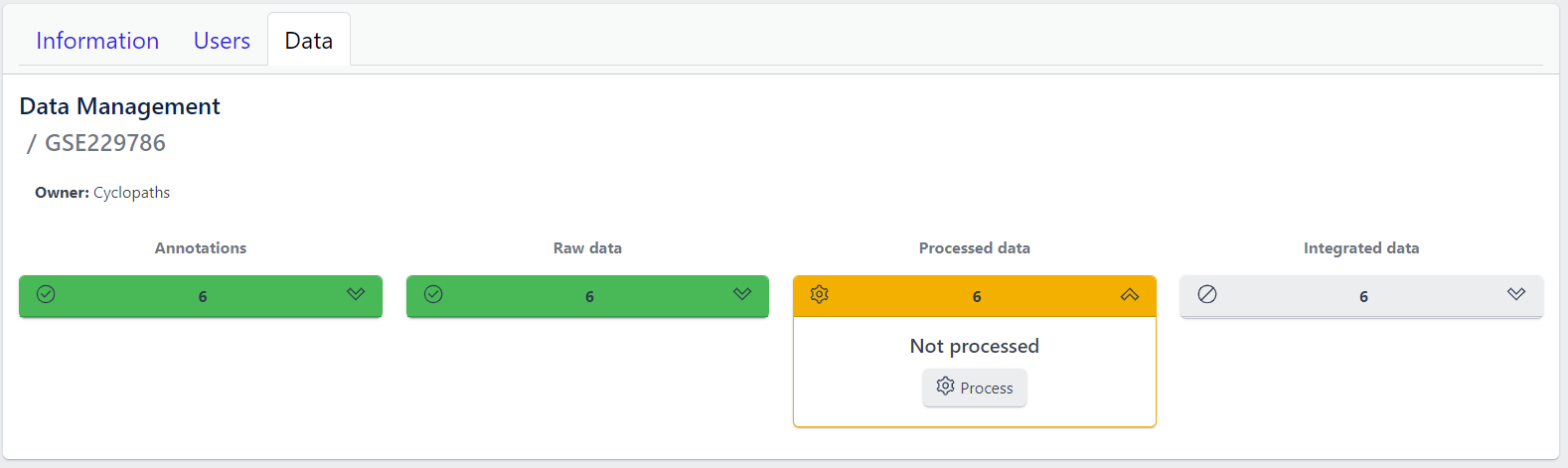
Step 2: Click on the Process button
Once the processing is initiated, a background job is started on the server. Blue coloured Processed data indicates that the job is running. Please keep in mind that depending on dataset sizes, these jobs may run for multiple hours or even days.
Once sucessfully completed, the Processed data stage turns green, displaying Processed status. Additionally, Integrated data tile is now coloured yellow, indicating that the data is ready for integration.
In case of a failure, Processed data stage turns red displaying Error status. To investigate what went wrong, please click on the View button to see output of the job. Clicking on the Retry button will start data processing again.
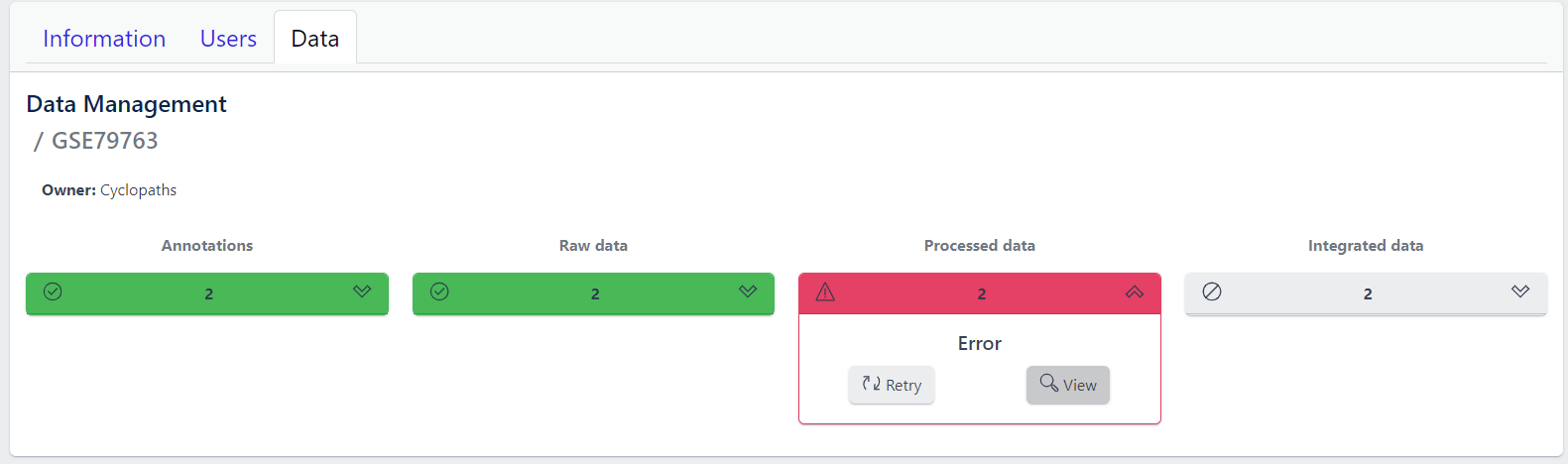
Once processed data is available in the study, the data is ready to be integrated to PanHunter. The pipeline is ready for integration once Processed data is coloured green and Integrated data is coloured yellow. Data will be available in PanHunter only after integration process is successfully finished.
Similar to the previous stages described above, the Integrated data tile in the study details view allows to run data integration jobs.
Step 1: Unfold Integrated data tile in the detailed study view.
Step 2: Click on the Integrate button
Clicking on the Integrate button starts a background job on the server. While the process is running, the Integrated data tile will be coloured blue.
Once sucessfully completed, the Integrated data stage turns green and data automatically becomes available in PanHunter apps.

In case of a failure, the Integrated data turns red displaying Error status. To investigate what went wrong, please click on the “View” button to see output of the job. Clicking on the Retry button will start data processing again.
Studies already created and existing in PanHunter, can be updated at any time via detailed study view. This can be done at any stage of data upload pipeline, regardless whether the study has been fully integrated into PanHunter or not.
In order to modify the sample metadata, to either correct or add additional information, it is possible to download and re-upload sample table files via the green Annotation tile in the detailed study view. This can also be done when the raw data have been added, processed and integrated already.

The process is similar to creating new samples as described in here, except that in this case it’s possible to start with sample table that is already available for the study.
Notes
In case update of data is required, this can be done for any stage at any time. To re-upload raw data or re-run processing and/or integration, please use Update button for respective data upload stage in the detailed study view.
Please note that updating raw or processed data requires re-integration of the data as well.
Deleting a study from PanHunter is possible directly in the overview of available studies in Data tab via Delete button.
Clicking on the Delete button will trigger a pop-up window to confirm deletion of the study.
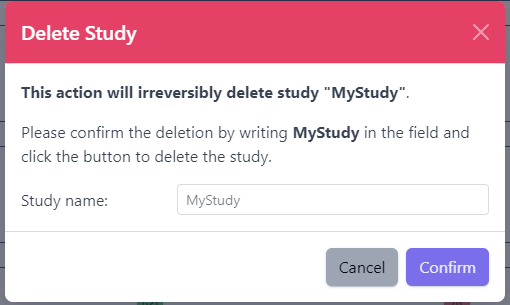
Please note that at the moment, possibility to delete study depends on the study data type (e.g. RNA seq data). In case you experience issues, please contact PanHunter Support.